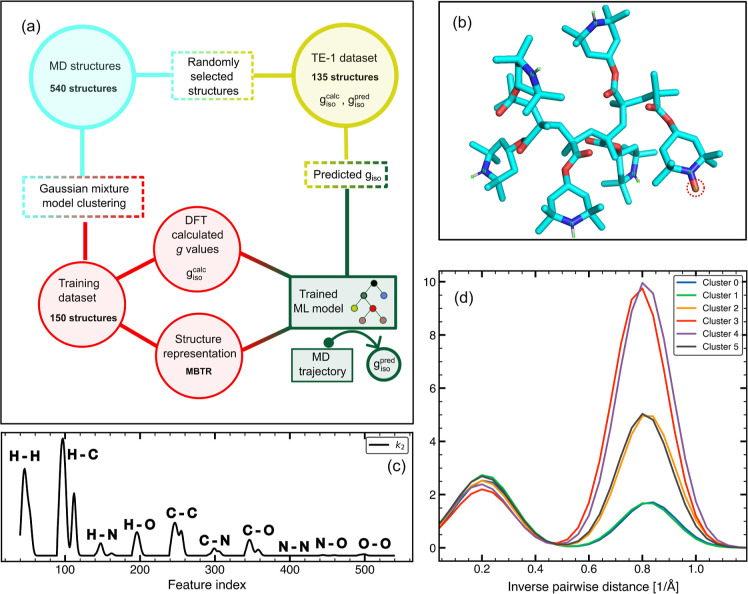
Figure 1.
(a) Schematic representation of the workflow used for machine learning giso. (b) Ball and stick model of PTMA-1, with six monomer units out of which one is a radical center (circled with a red dotted line). Oxygen atoms are colored red, nitrogen atoms are colored blue, and carbon atoms are colored cyan. (c) Schematic representation of the k2 term of MBTR for a PTMA-1 molecule. (d) Section of the N–O region of MBTR showing structural differences in cluster centers obtained using GMM. The distribution centered at an inverse pairwise distance of ≈0.8 Å–1 corresponds to the N–O bond length, and the distribution centered at ≈0.2 Å–1 corresponds to long-range N–O interactions. The distributions are weighted exponentially to give more weight to short-range interactions.