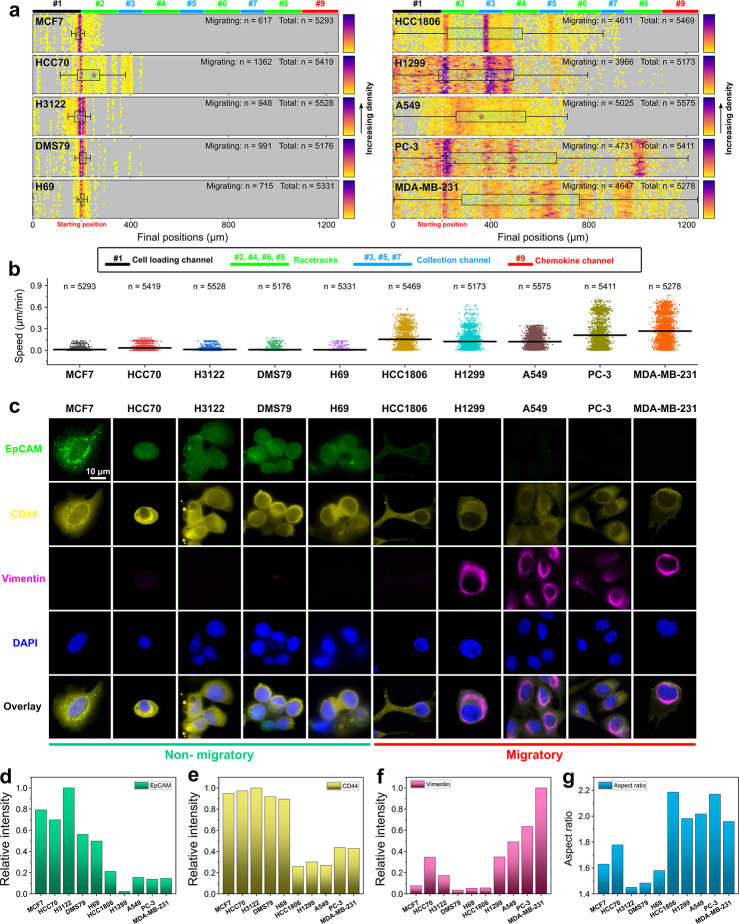
Figure 3.
Validation of CTC-Race assay using breast, lung, and prostate cancer cell lines. ∼5000 cancer cells were loaded into the device for each cell line experiment. Optimized conditions (0.1 μL/min perfusion rate, 10% FBS concentration, and 24-h assay time) were used for all experiments. After the assay, cells were fixed and stained with four markers (EpCAM, vimentin, CD44, and DAPI). Migration distance, migration speed, cellular aspect ratio, and marker expressions of cells were analyzed from bright-field and fluorescent imaging. (a) Plots of migration distances of 10 different cancer cell lines in the CTC-Race assay (non-small-cell lung cancer, A549, H3122, H1299; small-cell lung cancer, DMS79, H69; breast cancer, HCC70, HCC1806, MCF7, and MDA-MB-231; prostate cancer, PC-3). Box plots include the smallest, lower quartile, mean, upper quartile, and largest final positions for cells. Mean final positions are 191.53 ± 33.03 μm (MCF7, n = 5293), 232.09 ± 77.47 μm (HCC70, n = 5419), 190.57 ± 33.32 μm (H3122, n = 5528), 199.03 ± 30.05 μm (DMS79, n = 5176), 198.48 ± 24.43 μm (H69, n = 5331), 384.76 ± 195.81 μm (HCC1806, n = 5469), 318.25 ± 189.51 μm (H1299, n = 5173), 358.96 ± 143.91 μm (A549, n = 5575), 467.88 ± 317.74 μm (PC-3, n = 5411), and 556.89 ± 303.54 (MDA-MB-231, n = 5278). All values are mean ± sd. (b) Migration speed of cancer cell lines. Mean speed for each cell line: 0.01 ± 0.02 μm min–1 (MCF7, n = 5293), 0.03 ± 0.05 μm min–1 (HCC70, n = 5419), 0.01 ± 0.02 μm min–1 (H3122, n = 5528), 0.01 ± 0.02 μm min–1 (DMS79, n = 5176), 0.08 ± 0.02 μm min–1 (H69, n = 5331), 0.15 ± 0.11 μm min–1 (HCC1806, n = 5469), 0.12 ± 0.10 μm min–1 (H1299, n = 5173), 0.12 ± 0.09 μm min–1 (A549, n = 5575), 0.21 ± 0.20 μm min–1 (PC-3, n = 5411), and 0.27 ± 0.19 μm min–1 (MDA-MB-231, n = 5278). All values are mean ± sd. (c) Selected immunofluorescence images of migratory cells from cell lines. Four channels are used in the immunofluorescence staining, including an epithelial maker EpCAM (green), a mesenchymal marker vimentin (magenta), a cell-migration marker CD44 (yellow), and a nucleus marker DAPI (blue). (d–f) Mean intensities (n = 2000) of EpCAM, vimentin, and CD44 markers across cancer cell lines. The intensities are normalized using maximal intensity (EpCAM, H3122; vimentin, MDA-MB-231; CD44, H3122). (g) Mean cellular aspect ratio of cancer cell lines (n = 2000).