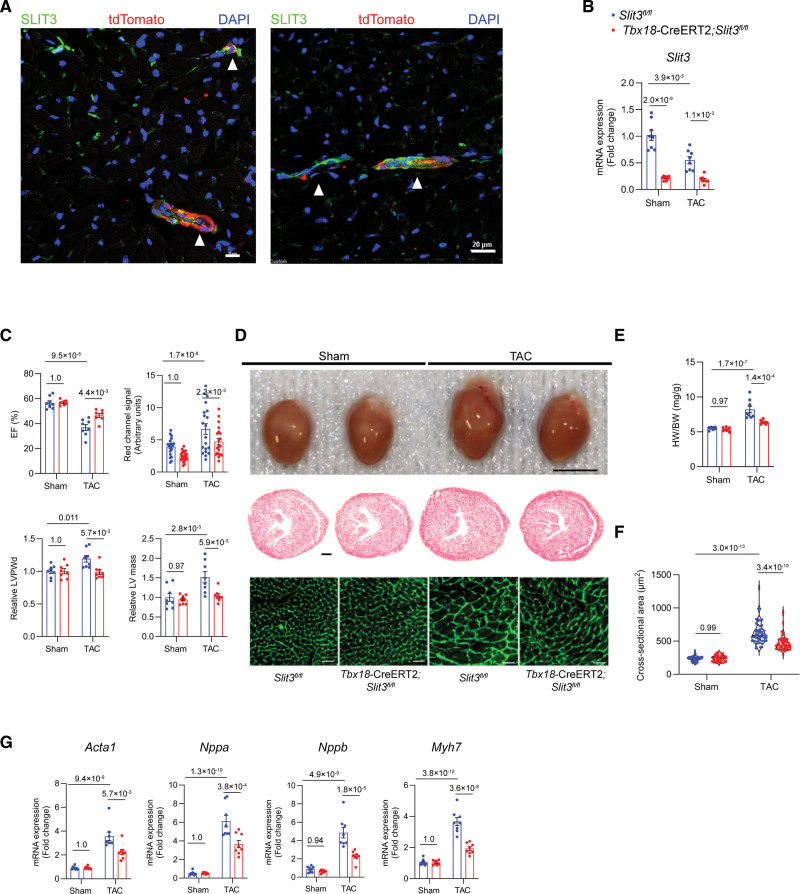
Figure 5.
Vascular mural cell–specific deletion of SLIT3 reduces cardiac hypertrophy and dysfunction after transverse aortic constriction (TAC). A, Immunofluorescence staining of heart sections from Tbx18-CreERT2;Rosa26-Tdtomato mice using anti-SLIT3 antibody (green), anti-tdTomato antibody (red), and 4’,6-diamidino-2-phenylindole (DAPI; blue). White arrowheads denote the overlap of SLIT3 and tdTomato signals in the mural region of myocardial blood vessels in cross-section (left) and axially (right). Scale bar for (left), 10 µm. Scale bar for (right), 20 µm. B, Slit3 transcription determined by quantitative PCR (qPCR; normalized to Gapdh transcript levels) in Slit3fl/fl or Tbx18-CreERT2;Slit3fl/fl mice hearts after sham or TAC surgery. N=8 mice/group. C, Echocardiographic studies of Slit3fl/fl and Tbx18-CreERT2;Slit3fl/fl mice after sham or TAC surgery. N=8 mice/group. D, Appearance of hearts explanted from Slit3fl/fl control and Tbx18-CreERT2;Slit3fl/fl mice after sham or TAC surgery (top); representative images of heart sections stained with hematoxylin & eosin (H&E; scale bar, 500 µm; middle); and representative images of heart sections stained with wheat germ agglutinin (WGA; scale bar, 20 µm; bottom). E, Heart weight to body weight (HW/BW) ratio. N=8 mice/group. F, Quantification of cardiomyocyte cross-sectional area on WGA staining. N=600 cells from 6 mice in each group. G, qPCR analysis of hypertrophy-associated gene transcription (Nppa, Nppb, Myh7, and Acta1, normalized to Gapdh mRNA expression) in hearts from Slit3fl/fl control or Tbx18-CreERT2, Slit3fl/fl mice after sham or TAC surgery. N=8 mice/group. Two-way ANOVA with the Tukey multiple comparisons test was used in all analyses. EF indicates ejection fraction; LV, left ventricle; and LVPWd, end-diastolic left ventricle posterior wall thickness.