Figure 2.
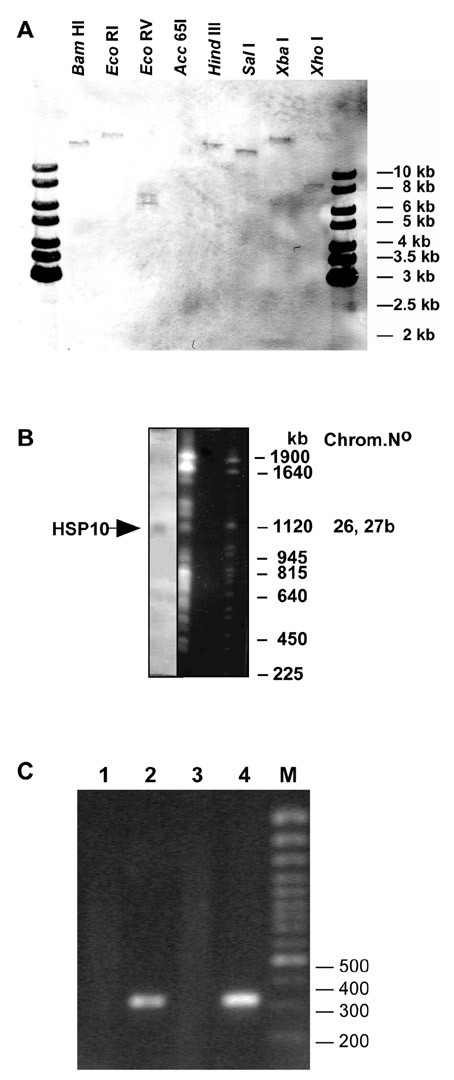
(A) Southern analysis of L. donovani genomic DNA. The DNA (2 μg) was digested with restriction enzymes as indicated and separated on a 1 % agarose gel. After Southern transfer the membrane was probed with a CPN10 specific probe. The positions of size markers (1 kb ladder, Fermentas) are indicated. (B) Pulsed field gel electrophoresis of L. donovani chromosomes. The gel was stained with ethidium bromide and photographed (right). After denaturation the DNA was blotted on a Nylon membrane and probed with a CPN10 specific probe (left). The sizes of S. cerevisiae chromosomes which were run alongside as size markers are shown to the right. The chromosome band which hybridised with the CPN10 probe is marked by an arrow. (C) RT-PCR of L. donovani RNA. Whole cell RNA was isolated from L. donovani cells and subjected to reverse transcription at 37°C for 60 min using an oligo-dT primer. An aliquot of the cDNA reaction mix was inactivated at 65°C for 10 min prior to incubation at 37°C. Either RNA (lane 1), the cDNA (lane 2), the heat inactivated cDNA mix (lane 3) or genomic DNA (lane 4) were added to a PCR reaction mix using CPN10 specific primers. The sizes of selected marker DNA molecules is shown to the right.
