Fig. 2. Overstretch tension sensor.
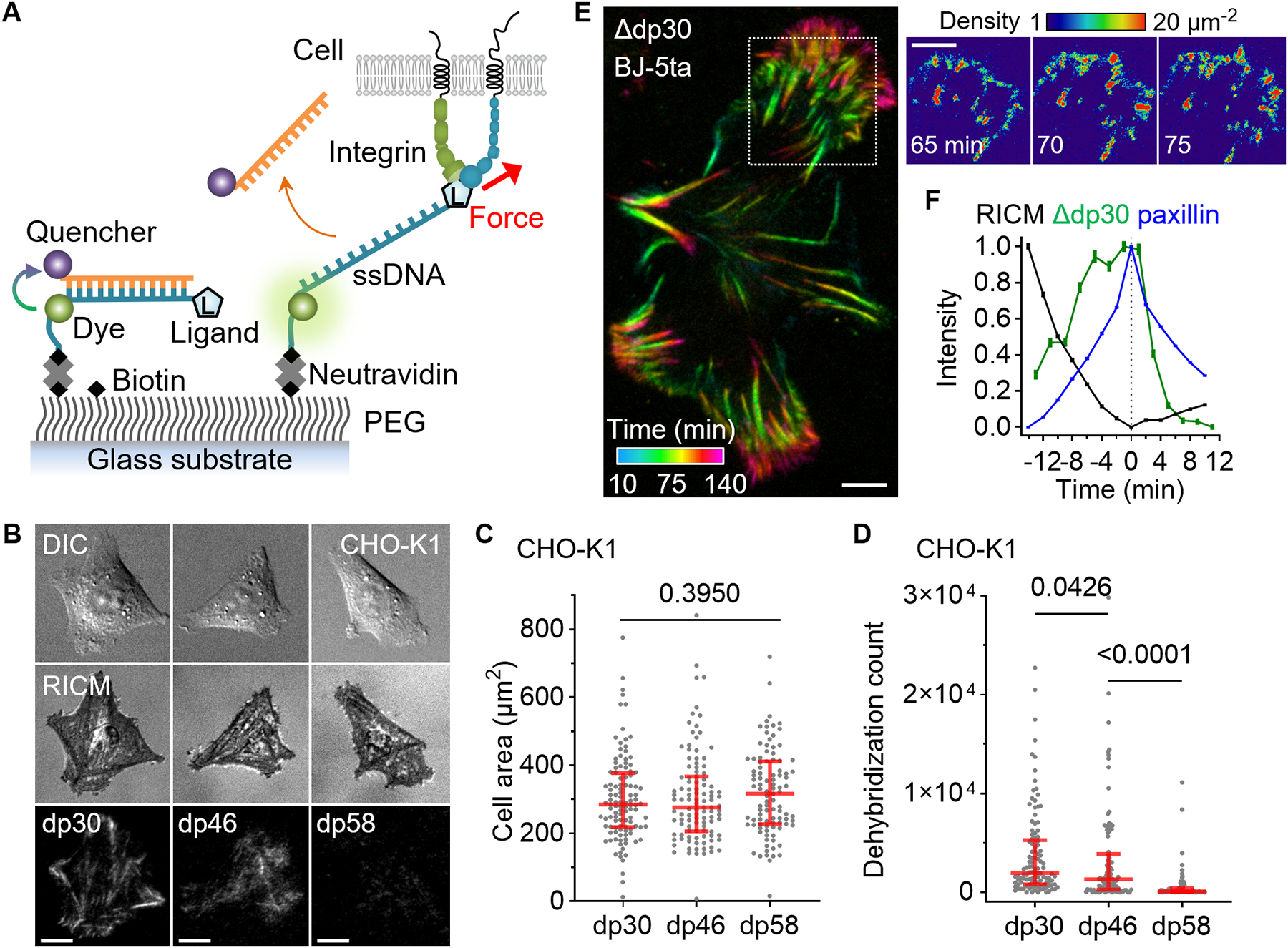
Single-molecular force sensors were created based on force-induced oligonucleotide dehybridization. (A) Schematic of force detection using OTS. (B) Images of epithelial-like CHO-K1 cells seeded (2 hours) on densely immobilized (~1000 μm−2) dp30, dp46, and dp58 conjugated with a cRGDfK ligand. (C) CHO-K1 cell area on the three OTS coated surfaces. N = 112, 109, and 99 cells from three independent experiments. (D) The count of dehybridized OTS per cell. The red lines are median and interquartile range. P-values are from the Kruskal-Wallis test. (E) BJ-5ta fibroblast force-activated dp30 signal is temporally resolved at 5 min intervals. The density of force events quantified by measuring single Cy3 signals. (F) Temporal relation between RICM, dp30 signal increase, and paxillin-GFP signals. Pixel-wise signal traces were aligned to the peak of GFP signal and averaged (2830 pixels; mean±SE). BJ-5ta fibroblast spreading was monitored at 2 min time intervals. Scale bars, 10 μm.
