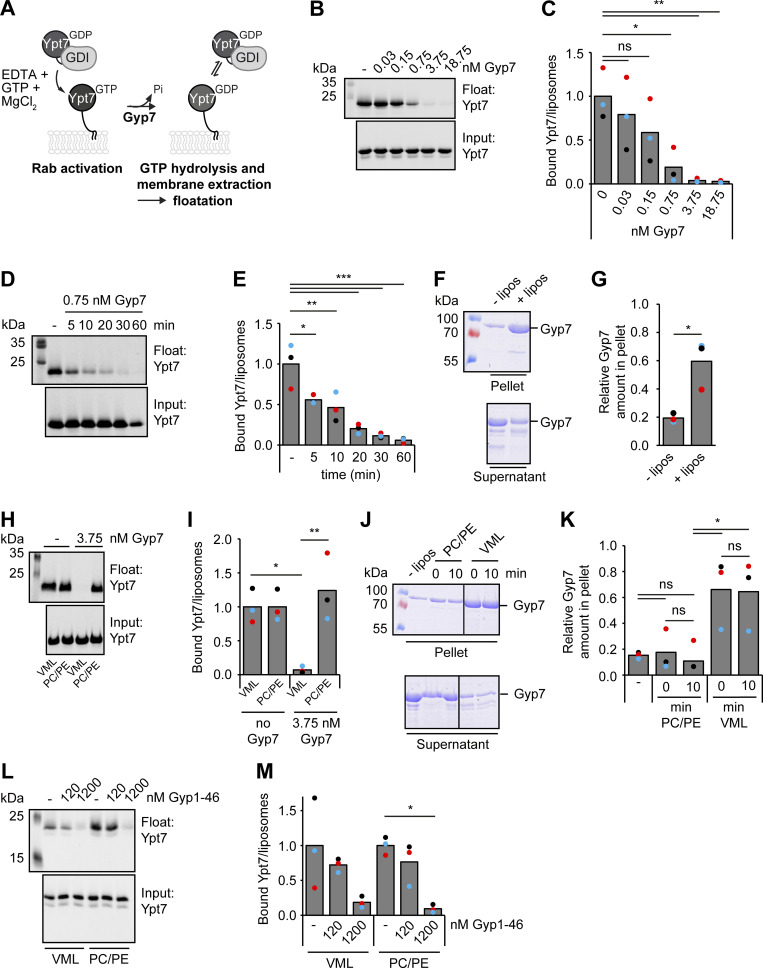
Figure 4.
Gyp7 requires a distinct membrane environment for efficient GAP activity. (A) Overview of the GDI extraction assay. 250 μM liposomes with VML composition are preloaded with 0.6 μM Ypt7-GDI complex in the presence of 3.75 mM EDTA and 125 μM GTP. The nucleotide binding is stabilized by addition of 7.5 mM MgCl2. Incubation with the GAP Gyp7 triggers GTP hydrolysis. GDI extracts inactivated Ypt7 from liposomal membranes. Liposomes with bound Ypt7 are floated in a sucrose gradient and separated from unbound protein. Floated membrane fractions and inputs were analyzed by western blotting (see Materials and methods). (B) Ypt7 inactivation increases with the concentration of Gyp7. Assay was performed as in A. Reactions were incubated with different amounts of Gyp7 for 1 h. Control reaction contained no Gyp7. 40% of the float was analyzed together with 3% input by western blotting using an anti-Ypt7 antibody. (C) Quantification of bound Ypt7 to liposomes in B. Band intensity of Ypt7 signal in float was measured in Fiji and compared with input. Reactions containing Gyp7 were normalized to the average value of the control reaction. Bar graphs represent the averages from three independent experiments and puncta represent the mean of each experiment. P value ns, *<0.05, **<0.01, using ANOVA one-way test. (D) Kinetics of Gyp7 activity toward Ypt7-GTP. Assay was performed as in A. Reactions were incubated with 0.75 nM Gyp7 for different time points. Control reaction contained no Gyp7. 40% of the float was analyzed together with 3% input by western blotting using an anti-Ypt7 antibody. (E) Quantification of bound Ypt7 to liposomes in D. Quantification was performed as in C. P value *<0.05, **<0.01, ***<0.001 using ANOVA one-way test. (F) Membrane association of Gyp7. 715 μM liposomes with VML composition were incubated with 715 nM Gyp7 for 10 min. Membranes were separated from supernatant by centrifugation at 100,000 g and both fractions were analyzed by SDS-PAGE and Coomassie staining. Control reaction contained no liposomes (see Materials and methods). (G) Quantification of the relative Gyp7 amount in the pellet in F. Band intensity of Gyp7 signal in the pellet was measured in Fiji and compared with Gyp7 signal in the supernatant. Bar graphs represent the averages from three independent experiments and puncta represent the mean of each experiment. P value *<0.05, using two-sided Student’s t test. (H) Comparison of Gyp7 activity on liposomes with VML composition and PC/PE liposomes. The assay was performed as in A. 3.75 nM Gyp7 was added to reactions containing liposomes with VML composition or PC/PE liposomes for 10 min. Control reactions contained respective liposomes and no Gyp7. 40% of the float was analyzed together with 3% input by western blotting using an anti-Ypt7 antibody. (I) Quantification of bound Ypt7 to liposomes in H. Quantification was performed as in C. Reactions containing Gyp7 were normalized to the average value of the respective control reaction. P value *<0.05, **<0.01, using ANOVA one-way test. (J) Association of Gyp7 with liposomes of VML composition and PC/PE liposomes. 715 nM Gyp7 was incubated with 715 μM liposomes for 0 and 10 min. Membrane association was analyzed as in F. (K) Quantification of the relative Gyp7 amount in the pellet in J. Quantification was performed as in G. P value ns, *<0.05, using ANOVA one-way test. (L) Comparison of Gyp1-46 activity on liposomes with VML composition and PC/PE liposomes. Assay was performed as in A, except for the addition of Gyp1-46 instead of Gyp7 to reactions. Reactions were incubated with different amounts of Gyp1-46 for 10 min. Control reactions contained respective liposomes and no GAP. 40% of the float was analyzed together with 3% input by western blotting using an anti-Ypt7 antibody. (M) Quantification of bound Ypt7 to liposomes in L. Quantification was performed as in C. Reactions containing Gyp1-46 were normalized to the average value of the respective control reaction. P value *<0.05, using ANOVA one-way test. Source data are available for this figure: SourceData F4.