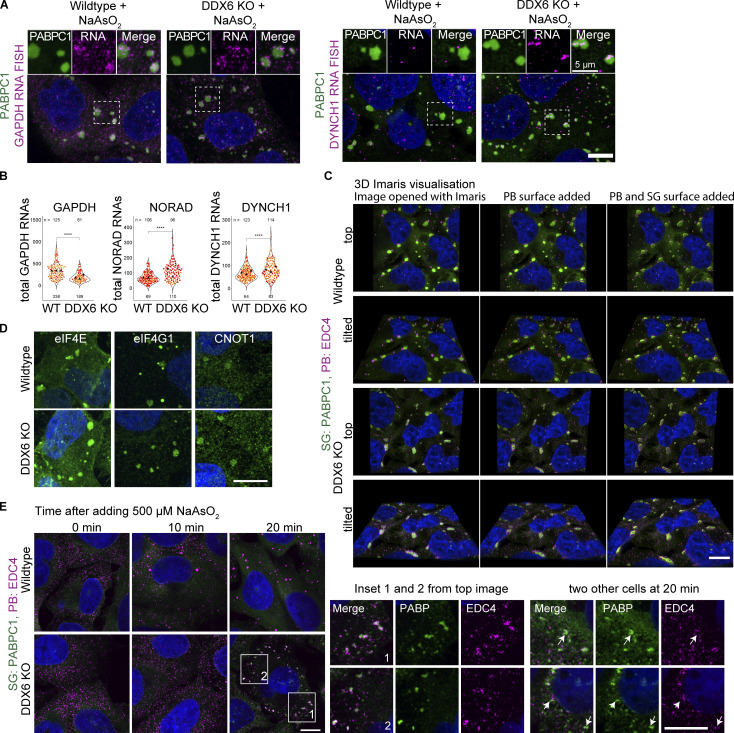
Figure S3.
Supporting data for increased RNA enrichment and the formation of smaller PB like assemblies in DDX6 KO cells. (A) IF images comparing GAPDH and DYNCH1 RNA localization in SGs in wild-type and DDX6 KO cells. (B). Quantification of the total number of GAPDH, DYNCH1, and NORAD RNAs in the cytoplasm. For quantification, data points from three biological replicates (two for NORAD) are shown in red-orange-pink, the mean replicate values are indicated as triangles, and mean values are shown at the bottom of the image (m). ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05, ns P > 0.05 (unpaired, two-tailed t test on individual data points). (C) Imaris 3D visualization in wild-type (top) and DDX6 KO (bottom) cells. Z-stacks are opened with Imaris (left) and PB surfaces (center) or PB and SG surfaces (right) are added. (D) IF images showing eIF4E, eIF4G, and CNOT1 localize to both granules in wild-type cells or in SGs in DDX6 KO cells. (E) Top: IF images showing the increase in SGs and PBs/PB-like assemblies (EDC4 IF, magenta; PABPC1, green) in wild-type and DDX6 KO cells at 10 and 20 min of stress treatment, to highlight the formation of interacting PB-like assemblies and smaller SGs in DDX6 KO cells. EDC4 IF signal is overamplified to highlight the lack of PB-like assemblies at earlier timepoints and in cells without SGs. White boxes denote the insets at the bottom. Bottom: Close-up view of the two indicated insets and two additional close-up views at 20 min of other cells with less or smaller PB-like assemblies.