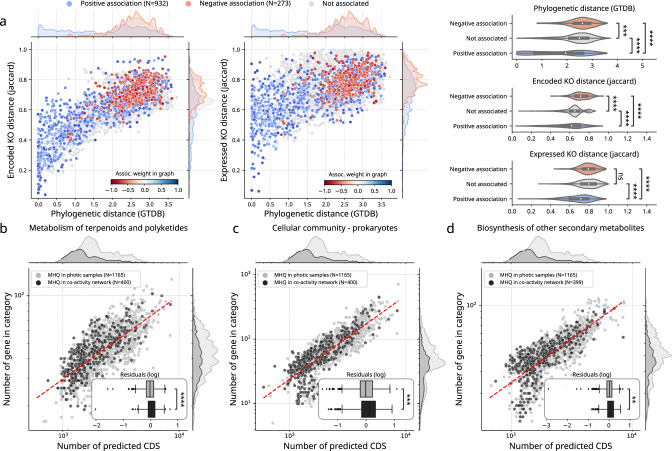
Fig. 3. A genome-resolved co-activity network reveals biotic factors shaping marine prokaryotic community structure.
a A genome-resolved co-activity network was inferred from genome-wide activities in euphotic samples, and revealed a larger number of significant positive associations between genomes across diverse phylogenetic distances. Based on encoded KO gene presence/absence, and expressed KO gene presence/absence, using Jaccard distances between genomes as a proxy for functional distance (using KEGG), co-active genomes were functionally closer than expected at random (p = 1.187 × 10 − 25, two-sided Mann–Whitney U test on log-transformed distributions). b–d Scaling laws in the functional content of genomes highlighted broad metabolic categories enriched in co-active genomes versus genomes detected as active in samples. Notably, co-active genomes displayed a higher functional potential for terpenoid and polyketide metabolism (p = 1.46 × 10 − 7, two-sided Mann–Whitney U test on log-transformed distributions), for cellular community metabolism (quorum-sensing and biofilm formation, p = 4.00 × 10 − 4, two-sided Mann–Whitney U test on log-transformed distributions), and for the biosynthesis of other secondary metabolites (p = 4.73 × 10 − 9, two-sided Mann–Whitney U test on log-transformed distributions). Dashed-red lines are the best linear fit on a log-log scale (parameters given in Supplementary Data 3). The box extends from the lower to upper quartile values of the data (Q1 and Q3), with a line at the median (Q2). The whiskers extend from the box to show the range of the data and are defined as follows: where IQR is the interquartile range (Q3-Q1), the upper whisker will extend to last data point less than Q3 + 1.5 × IQR. Similarly, the lower whisker will extend to the first data point greater than Q1–1.5 × IQR. Beyond the whiskers, data are plotted as individual points. See Supplementary Data 6 for a complete list of functions enriched in co-active genomes.