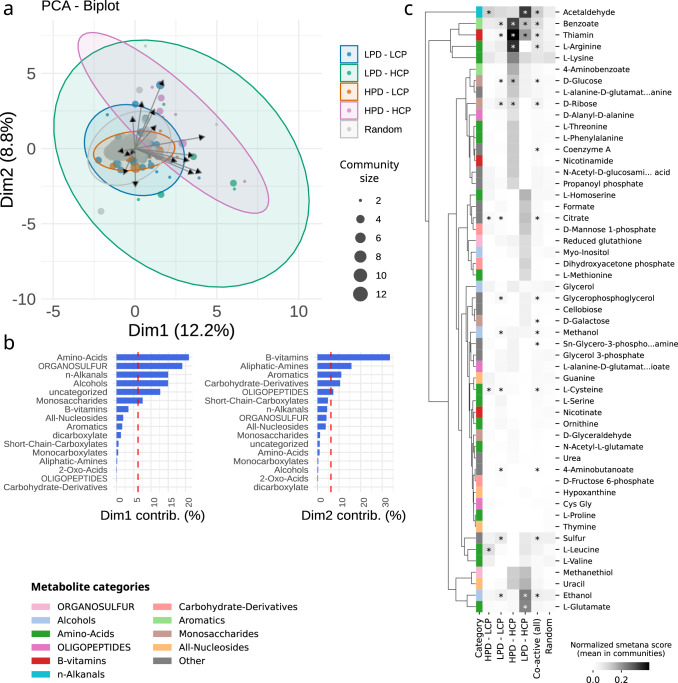
Fig. 5. Community metabolic modelling predicts specific metabolic cross-feedings within co-active marine prokaryotic communities.
a A NMDS analysis revealed that HPD-HCP and LPD-HCP communities are predicted to have a higher potential exchange in specific metabolic categories. Point size represents the size of each community in number of genomes. Coloured ellipses are visual aids to emphasize the distribution of points by categories. b Overall, the higher potential for exchanges in HPD-HCP and LPD-HCP communities is driven by specific metabolic categories (NMDS Dim1 and Dim2), in particular amino acids, B vitamins, organo-sulphur compounds, and aliphatic amines. c Within these broad metabolic categories, specific metabolite exchanges are identified within each co-active genome community type. Similar rows in the heatmap were clustered together by hierarchical clustering (UPGMA algorithm). In particular, exchanges of acetaldehyde, benzoate, thiamin (vitamin B1), ethanol, and L-glutamate are predicted in LPD-HCP, while in HPD-HCP exchanges of benzoate, thiamin, L-arginine, as well as D-glucose and D-ribose are predicted. Truncated metabolite names are (from top to bottom): L-alanine-D-glutamate-meso-2,6-diaminoheptanedioate-D-alanine, N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid, Sn-Glycero-3-phosphoethanolamine, and L-alanine-D-glutamate-meso-2,6-diaminoheptanedioate. Stars denote a significant difference between categories (Mann–Whitney U, Benjamini-Hochberg correction, corrected p value ≤ 0.05, all test results are available in Supplementary Data 8).