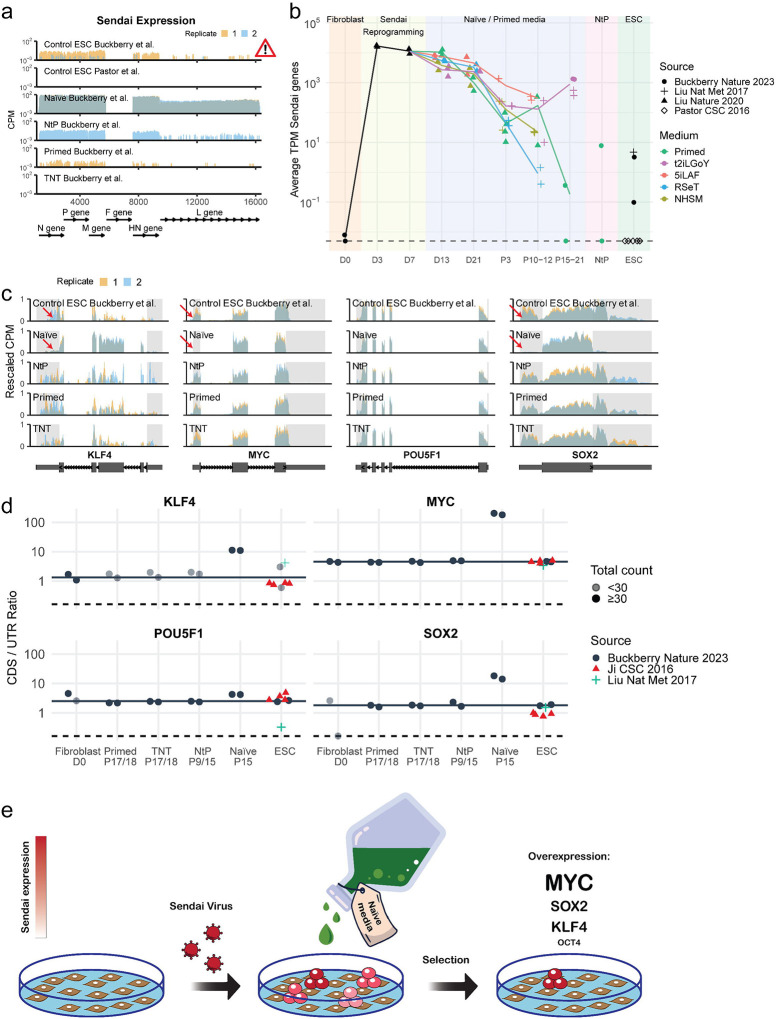
Fig. 1: Persistence of Sendai virus and selection for exogenous factor expression by naive media.
a, Read coverage of the Sendai virus genome. Two replicates per condition are shown (in red and blue). Coverage was binned at 100 bp resolution and is shown as counts per million reads (CPM). The F gene was deleted in the commercial kit used for reprogramming in these studies. b, Time-course of Sendai expression across reprogramming and hPSC samples. The y-axis shows the mean expression as transcripts per million (TPM) of all Sendai genes – except for F. Symbols indicate the study from which samples were sourced and colors indicate the cell culture media that the samples were grown in. The dashed line at the bottom corresponds to the location of samples with zero TPM. c, Read coverage of Yamanaka factors. The CPM values are rescaled such that each sample has a range from exactly zero to one. Large boxes in the gene models indicate exons, small boxes correspond to 5’ and 3’ untranslated regions (UTRs), and lines with arrows indicate introns. UTRs are highlighted using gray shading. d, Quantification of the ratio between coding sequence and untranslated region expression (CDS/UTR ratio). For each Yamanaka factor we quantified the number of reads aligning to CDS and UTR regions. The ratio of normalized counts CDS/UTR serves as a proxy for the proportion of transcripts originating from the viral transgene vs the endogenous locus. The horizontal black line is drawn at the mean ratio of the hESC control samples from Buckberry et al., which serve as baseline for the expected ratio in the absence of a transgene. The horizontal dashed line is drawn at the location of samples with a ratio of zero. Samples with low expression below <30 raw counts are drawn in a lighter shade. e, Graphical overview and mechanism of TNT method: the study utilizes t2iLGoY naive media, which preferentially selects for cells expressing high levels of exogenous reprogramming factors from Sendai virus RNA (illustrated in dark red). This selective expression of exogenous factors may significantly contribute to the outcomes observed in the TNT reprogramming.