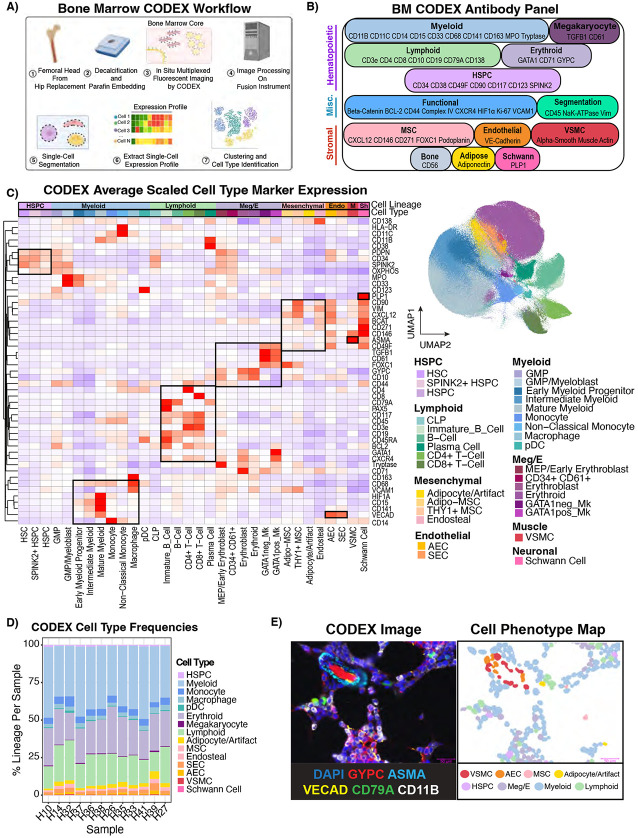
Figure 4 – 54-plex CODEX imaging reveals the spatial cellular topography of human bone marrow.
A) Schematic depicting the CODEX experimental and computational workflow leading to cell type identification. B) Diagram showing the 54-plex CODEX panel (53 antibodies + DAPI) split by target cell population. C) Heatmap showing average centered-log-odds ratio normalized expression per cell type scaled and clustered by protein showing the marker protein expression is consistent with literature knowledge (Left). Cell types were annotated by unsupervised clustering and manual gating as described in Materials and Methods. Boxes were manually drawn to highlight coordinate marker expression. UMAP showing the 803,131 single cells in the CODEX atlas from 12 individuals colored by cell type (Right). Adipocytes were labeled with /Artifact because this population contained a mixture of true adipocytes and artifactual staining. HSPC – Hematopoietic Stem and Progenitor Cell. Meg/E-Megakaryocyte/Erythroid, Endo-Endothelial, M-Vascular Smooth Muscle, Sh-Schwann Cells. D) Stacked bar plot showing cell type frequencies normalized by total cells per sample. E) CODEX image (left) is paired with the cell phenotype map (CPM,right). An arteriolar structure and hematopoietic cells are shown using selected relevant fluorescent markers, which is juxtaposed with the same image with the cellular segmentation masks colored by cell type showing how CODEX allows single-cell mapping of the bone marrow microenvironment.