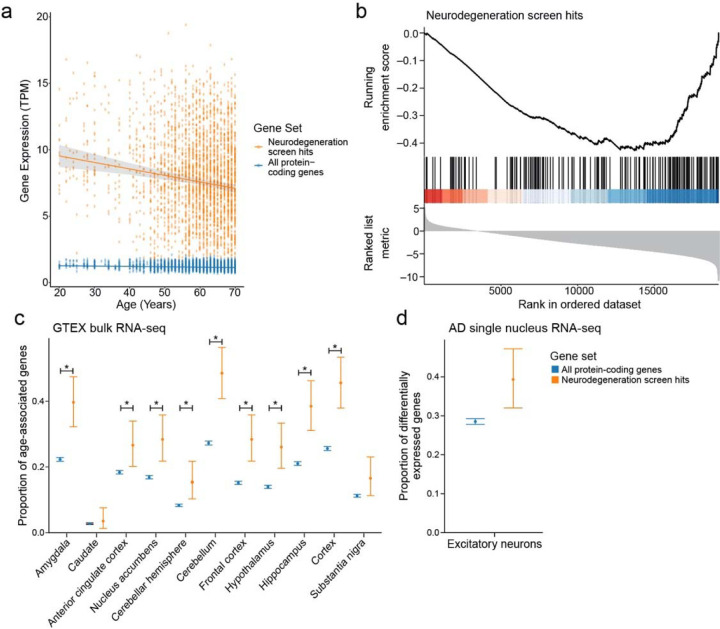
Fig. 2:
a) Geometric mean expression in transcripts per million (TPM) of neurodegeneration screen hits (neurodegeneration genes, orange) and all protein-coding genes in the Genotype-Tissue Expression (GTEx) shows that the expression of neurodegeneration screen hits declines with age in human brain tissues (all protein-coding genes: p=2.91*10−4, neurodegeneration screen hits: p=1.14*10−5). There is a significant difference in the slopes of the trends between age and gene expression for neurodegeneration screen hits and all protein-coding genes (all protein-coding genes: R=0.12, neurodegeneration screen hits: R=0.15, p=7.38*10−6). Regression lines indicate the relationship between age and TPM with a 95% confidence interval (standard error of the mean). The mixed effects regression analysis controlled for post-mortem interval, sex, ethnicity, and tissue of origin. b) Gene set enrichment plot showing that the set of age-associated neurodegeneration genes has reduced expression with respect to age. Vertical lines indicate rank of neurodegeneration screen hits by their association between gene expression and age determined by mixed-effects regression analysis coefficients. c) Proportion of genes that have significant associations between gene expression and age relative to the set of all protein-coding genes (blue) or the set of age-associated neurodegeneration genes (orange). Error bars indicate 95% binomial confidence intervals of the estimated proportion of genes with a significant association with age. Asterisk indicates tissues with an FDR-adjusted one-tailed hypergeometric test p-value less than 0.01. d) Proportion of protein-coding genes (blue) and age-associated neurodegeneration genes (orange) that are differentially expressed between Alzheimer’s disease (AD) and control in excitatory neurons in single-nucleus RNA-seq. Error bars indicate 95% binomial confidence intervals.