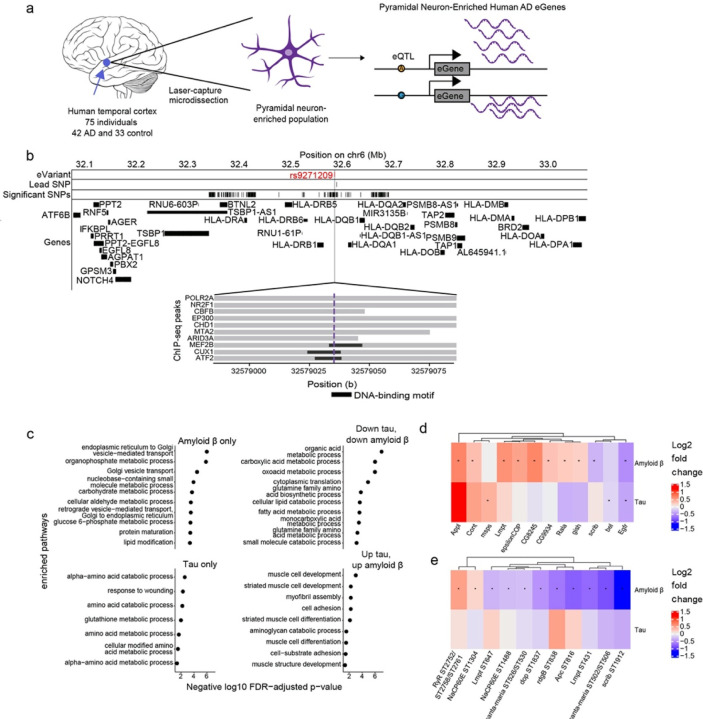
Fig. 3:
Multi-omic changes in human Alzheimer’s disease patients and model systems. a) Schematic depicting the identification of eGenes from laser-capture microdissection of temporal cortex pyramidal neuron-enriched populations from 75 individuals including 42 human Alzheimer’s disease (AD) and 33 healthy control patients and identification of eGenes. Brain cartoon created with Biorender.com. b) The eQTL associated with the eGene HLA-DRB1 is highlighted in red and overlaps with DNA binding motifs of MEF2B, CUX1 and ATF2 derived from ENCODE ChIP-seq and FIMO-detected motifs. Grey horizontal bars indicate ChIP-seq binding regions and the black horizontal bars indicate where the DNA-binding motif is located. c) Dot plots showing the negative log10 FDR-adjusted p-values for enriched GO terms in proteins that are significantly upregulated or significantly downregulated in both Drosophila models of tau and amyloid β, only differentially abundant in Drosophila models of amyloid β (Amyloid β only), or only differentially abundant in Drosophila models of tau (Tau only). d) Heat maps depict the log2 fold changes between Aβ1–42 transgenic flies (Amyloid β) or tauR406W (Tau) transgenic flies with controls for d) proteins or e) phosphoproteins that were hits in the age-associated neurodegeneration screen. An asterisk indicates whether the comparison was significant at an FDR threshold of 0.1. The columns of all heatmaps were clustered by hierarchical clustering.