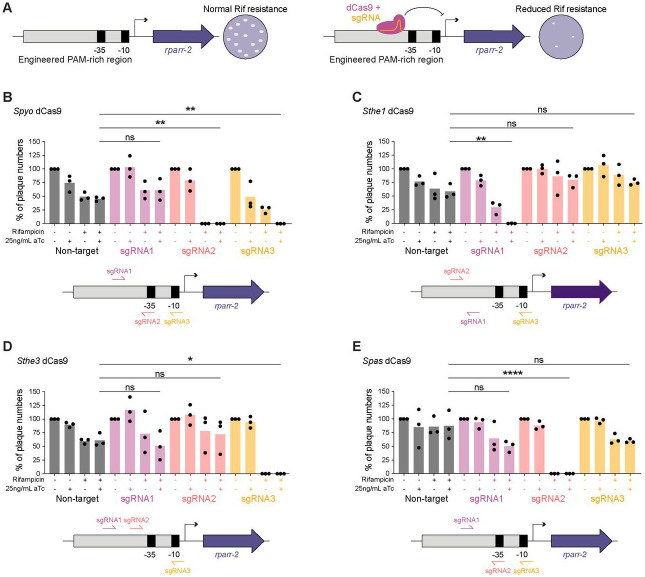
Figure 3. CRISPRi knockdown of a rifampicin resistance gene.
(A) Schematic of the engineered locus and screen to test knockdown of rifampicin resistance. The rpsL promoter driving expression of rparr-2 in the pMW1650 plasmid was modified to include additional PAMs to allow for testing of various dCas9 variants. Successful CRISPRi-mediated knockdown rparr-2 would sensitize strains to treatment with rifampicin, while strains with nonfunctional CRISPRi would remain resistant to rifampicin. Spectinomycin selection ensures that the strains maintain the plasmid encoding the CRISPRi components.
(B-E) Quantification of CRISPRi-mediated knockdown of rifampicin resistance via plaque assay. A549 cell monolayers were infected with R. parkeri strains encoding the S. pyogenes dCas9 (B), S. thermophilus 01 dCas9 (C), S. thermophilus 03 dCas9 (D), S. pasteurianus dCas9 (E). For each dCas9 variant and sgRNA combination, the same volume of R. parkeri stock was added to each well, and then the number of plaques was normalized to the no aTc and no rifampicin condition for a total of n = 3 independent experiments. Statistical significance was determined by ordinary one-way ANOVA with post hoc Tukey’s test (* denotes p < 0.05, ** denotes p < 0.005, **** denotes p < 0.0001). Schematics below each bar graph depict the relative locations of each sgRNA tested for each dCas9.