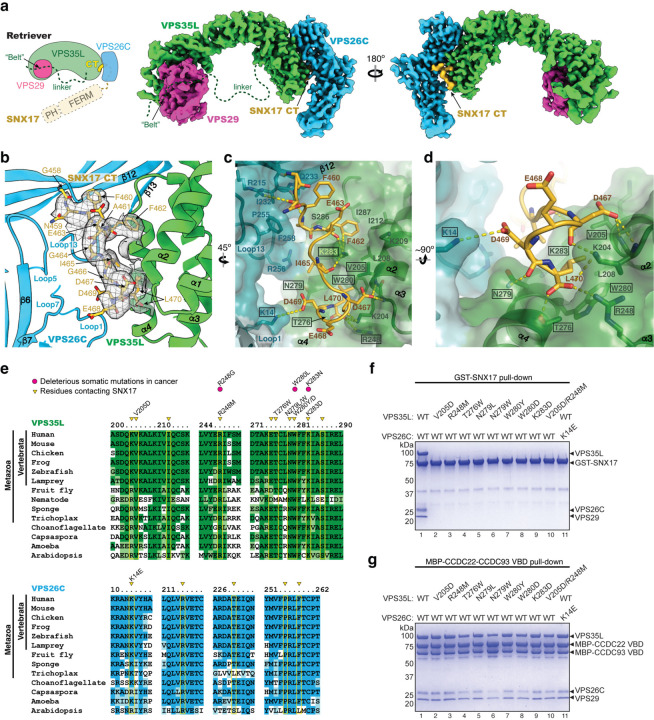
Fig. 2. Structure of SNX17 C-terminal tail binding to Retriever.
a. Schematic and overall colored cryo-EM map of the SNX17 CT peptide (golden) complexed with Retriever (VPS35L in green, VPS29 in magenta, and VPS26C in cyan). Dotted lines indicate structural elements not resolved in cryo-EM. N-terminal domains of SNX17 are shown as a reference. b-d. Close-up views showing key interactions between the SNX17 CT peptide (carbon in green, oxygen in red, and nitrogen in blue) and its binding surface on VPS35L and VPS26C: (b) shows cryo-EM density of the SNX17 CT peptide; (c-d) shows surface conservation calculated with ConSurf33, with color-to-white gradients representing the most (ConSurf score = 9) to the least conserved residues (ConSurf score = 1). Contacting residues are shown as sticks. Dotted yellow lines indicate polar interactions. Residues mutated in this study are indicated by a black box. e. Sequence alignment of human VPS35L and VPS26C with orthologs from indicated representative species. Residues contacting SNX17 are indicated with yellow boxes and arrowheads. Deleterious somatic mutations found in the COSMIC database and mutations tested for binding to SNX17 are indicated. f-g. Coomassie blue-stained SDS PAGE gels showing GST-SNX17 (f) or MBP-CCDC22-CCDC93 VBD dimer (g) pulling down purified Retriever bearing the indicated point mutations in VPS35L or VPS26C. Representative results from at least two independent experiments are shown.