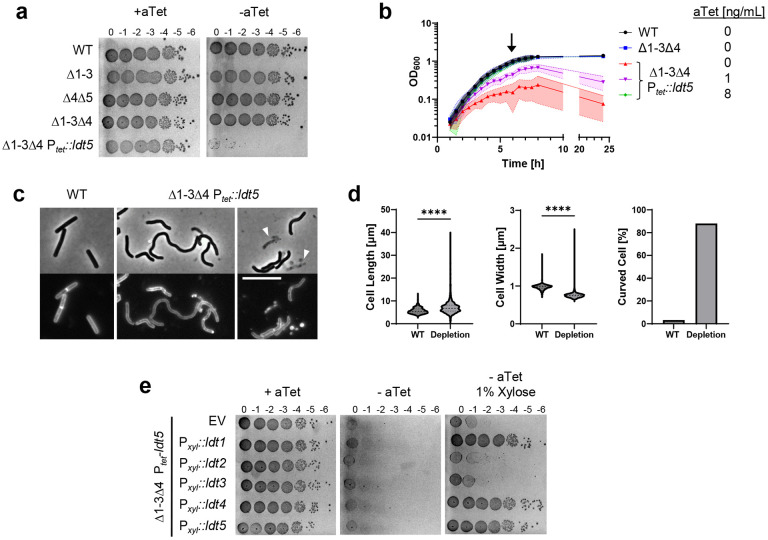
Fig. 4. LDTs are essential in C. difficile.
(a) Viability assay. Ten-fold serial dilutions of the indicated strains were spotted onto TY plates with or without 25 ng/mL aTet. Plates were photographed after incubation for 18 h. Images are representative of at least three experiments. (b) Growth curves. Data are graphed as the mean ± s.d. of 4 biological replicates from different days. (c) Cell morphology. Cells grown for 6 h in TY without aTet (arrow in B) were stained with the membrane dye FM4-64 and photographed under phase contrast and fluorescence microscopy. Arrowheads indicate lysed cells. Size bar, 10 μm. Images representative of at least three experiments. (d) Quantification of length, width and shape based on 781 cells of WT and 1196 cells of the depletion strain pooled from three biological replicates. Cells with a sinuosity score ≥1.03 were considered curved. ****, p < 0.0001, unpaired t-test. (e) Complementation assay. Tenfold serial dilutions of the LDT depletion strain harboring the indicated expression plasmids were spotted onto TY with or without 25 ng/mL aTet and 1% xylose. Plates were photographed after incubation for 18 h. Images are representative of three biological replicates. Strains shown in A-D: WT, R20291; Δ1-3, KB124; Δ4Δ5, KB529; Δ1-3Δ4, KB474;and Δ1-3Δ4 Ptet::ldt5, KB547 (called “depletion” in panel d). Strains shown in panel e: empty vector (EV), KB548, Pxyl::ldt1, KB549; Pxyl::ldt2, KB550; Pxyl::ldt3, KB551; Pxyl::ldt4, KB552; and Pxyl::ldt5, KB553.