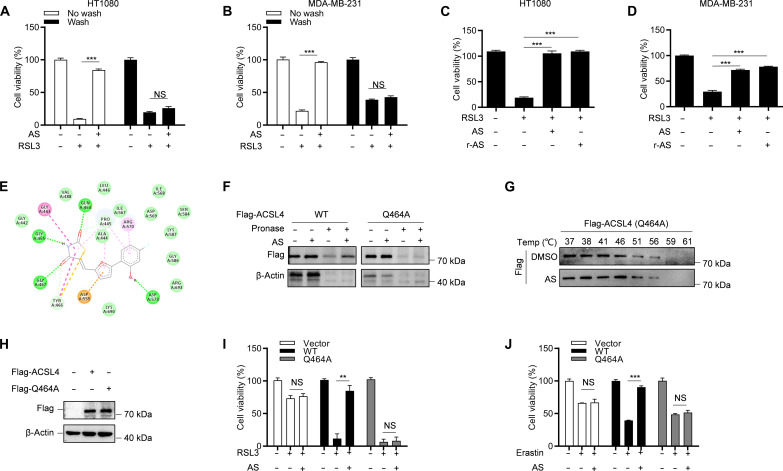
Fig. 4. AS binds to the Gln464 residue of ACSL4 to inhibit its activity.
(A and B) Cell viability analysis of HT-1080 cells (A) or MDA-MB-231 cells (B) pretreated with AS for 30 min and washed three times and then stimulated with 1 μM RSL3 for 8 hours. (C and D) Cell viability analysis of HT-1080 cells (C) or MDA-MB-231 cells (D) pretreated with AS or r-AS for 1 hour and then stimulated with 1 μM RSL3 for 8 hours. (E) Three-dimensional binding mode diagrams between ACSL4 and AS. (F) The cell lysates of HEK293T overexpressing WT or Q464A mutant Flag-ACSL4 were incubated with or without AS for 2 hours and then treated with different concentrations of pronase. The expression of ACSL4 was detected by immunoblotting. (G) The cell lysates of HEK293T overexpressing Flag-ACSL4 Q464A were incubated with or without AS for 2 hours and then treated with increasing melting temperature (37° to 61°C). The expression of ACSL4 was detected by immunoblotting. (H to J) CRISPR-Cas9–generated ACSL4-knockout HT-1080 cells were transduced with WT or Q464A mutant Flag-ACSL4 and then treated with AS for 1 hour and stimulated with RSL3 or erastin. The expression of ACSL4 was detected by immunoblotting (H), and the cell viability was analyzed by CCK-8 [(I) and (J)]. Data are the mean ± SEM, n = 3 biologically independent experiments [(A) to (D) and (I) and (J)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. NS, P > 0.05, **P < 0.01, ***P < 0.001.