Figure 4.
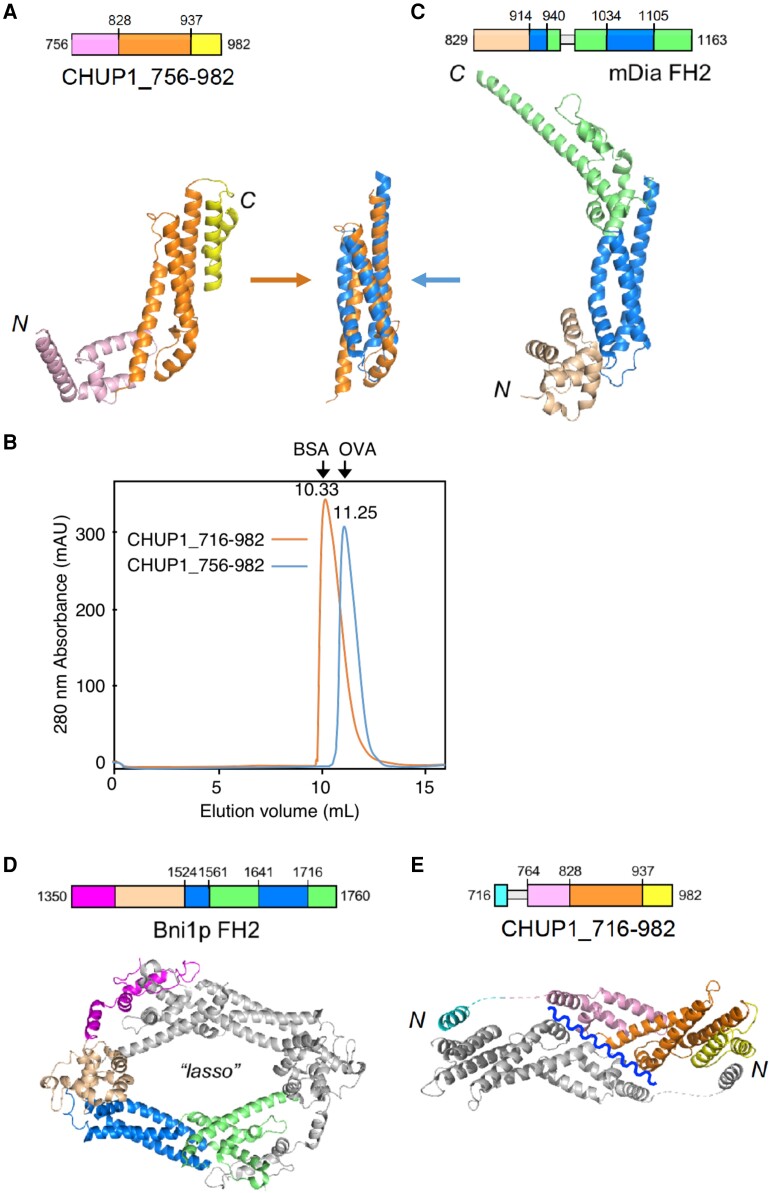
Three-dimensional structures of the C-terminal fragments of CHUP1. A) Crystal structure of CHUP1_756–982. B) The elution profiles of CHUP1_756–982 and CHUP1_716–982 were obtained by gel filtration chromatography using a Superdex 75 10/300 column. The elution volumes of BSA (67 kD) and ovalbumin (OVA, 43 kD) used for calibration are presented. The molecular weights of the recombinant proteins were calculated as 42 kD for CHUP1_756–982 and 64 kD for CHUP1_716–982. C) Crystal structure of the mDia1 FH2 domain (PDB 1V9D, Shimada et al. 2004). The central 3-helix bundles of CHUP1_756–982 and FH2 are superimposed in the middle between panels A and C). D) Functional dimer form of Bni1p FH2 (PDB 1UX5, Xu et al. 2004). E) Crystal structure of CHUP1_716–982. This fragment adopts a closed dimeric structure in crystal, which might open along the wavy line to become a functional dimer. In C and E), loops that are missing from the crystal structure due to poor electron density are indicated by thin gray boxes in the amino acid sequences.