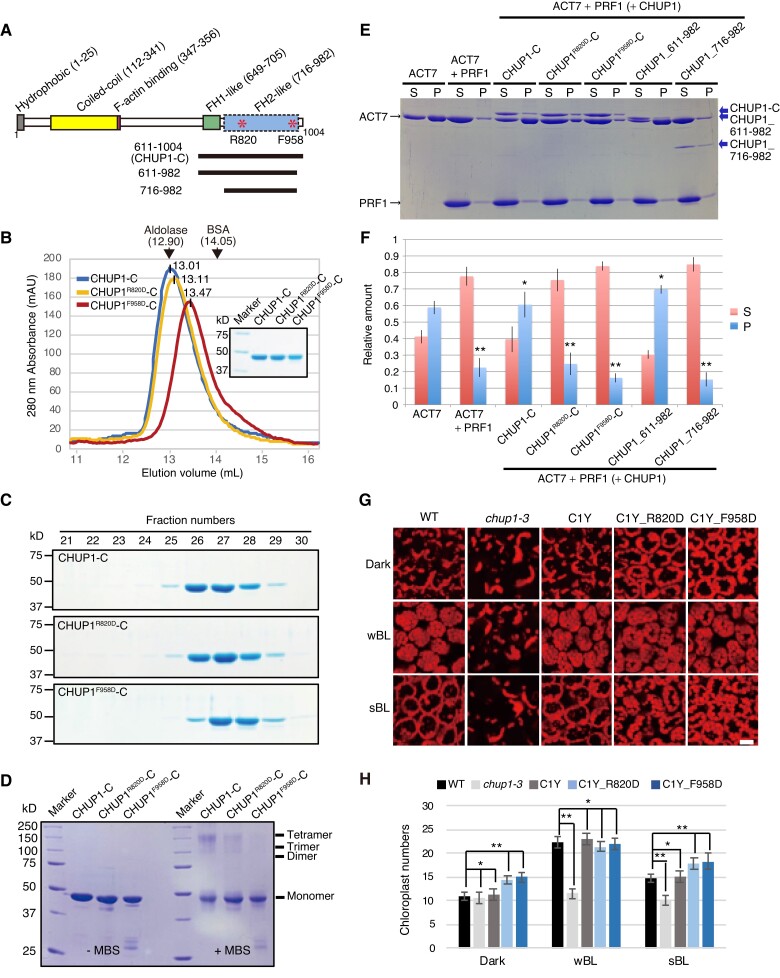
Figure 7.
Effects of point mutations at R820 or F958 of CHUP1-C on the structure and activity of CHUP1. A) Diagram of CHUP1 showing its functional domains: the hydrophobic region, the coiled-coil domain, the F-actin-binding domain, the proline-rich FH1-like domain, and the FH2-like domain. Asterisks indicate R820, which is involved in the association with actin, and F958, which is involved in the dimerization of the FH2-like domain. The positions of the 611 to 1,004 fragment (CHUP1-C) and the 611 to 982 and 716 to 982 fragments used for crystallization are indicated by lines below the structure. B and C) Effect of point mutations R820D and F958D on CHUP1-C dimer structure. B) The elution profiles of CHUP1-C, CHUP1R820D-C, and CHUP1F958D-C recombinant proteins were obtained by gel filtration chromatography using a Superdex 200 10/300 column. The elution volumes of aldolase (158 kD) and BSA (67 kD) used for calibration are presented. The molecular weights of the CHUP1-C recombinant proteins were calculated as 151.5 kD for CHUP1-C, 144.7 kD for CHUP1R820D-C, and 122.5 kD for CHUP1F958D-C. C) The protein profiles were confirmed using 10% SDS-PAGE gels. D) Effect of the point mutations R820D and F958D on the multimeric structure of CHUP1-C. Two µM of CHUP1-C, CHUP1R820D-C, and CHUP1F958D-C recombinant proteins was incubated without (left panel) or with (right panel) 20 µM MBS (m-maleimidobenzoyl-N-hydroxysuccinimide ester) for 1 h, and their multimer formations were analyzed using 10% SDS-PAGE. E and F) Effect of deletion and point mutations of CHUP1-C on actin polymerization. E) Ultracentrifugation assay of actin polymerization. ACT7 (4 µM) was allowed to polymerize in F-buffer 1 containing 8 µM PRF1 and 0.4 µM intact or mutant CHUP1-C, and a representative set of supernatant (S) and pellet (P) fractions after ultracentrifugation were analyzed by SDS-PAGE. F) Quantitative analysis of actin polymerization showing the fractions of ACT7 in the pellet and the supernatant under each condition as shown in E). The data are presented as means ± Sd (n = 3). Asterisks indicate statistically significant differences between ACT7 and each line of pellet detected by Student's t-test (* not significant P > 0.05; ** significant P < 0.0001). G) Effect of point mutations at R820D and F958D of CHUP1 on chloroplast positioning. The 4th rosette leaves of WT, chup1-3, C1Y, C1Y_R820D and C1Y_F958D plants were detached after dark adaptation (dark) for 14 h and then were further irradiated with a weak blue light of 2 µmol m−2 s−1 or a strong blue light (sBL) of 50 µmol m−2 s−1 for 2 h, respectively. Chloroplasts were shown with chlorophyll autofluorescence that was captured using confocal microscopy at a resolution of 512 × 512 pixels in a depth of 2 µm. Scale bar, 20 µm. H) Average number of chloroplasts observed on the periclinal side of palisade cells of WT, chup1-3, C1Y, C1Y_R820D, and C1Y_F958D plants. The average numbers of chloroplasts shown in G) were counted from 20 palisade cells from 3 rosette leaves. The data are presented as means ± SE (n = 20). Asterisks indicate statistical differences detected by Student's t-test (* not significant P > 0.05; ** significant P < 0.0001).