Background:
Nonalcoholic fatty liver disease (NAFLD) is a common chronic liver disease that imposes a huge economic burden on global public health. And the gut-liver axis theory supports the therapeutic role of intestinal flora in the development and progression of NAFLD. To this end, we designed bioinformatics study on the relationship between intestinal flora disorder and NAFLD, to explore the possible molecular mechanism of intestinal flora interfering with NAFLD.
Methods:
Differentially expressed genes for NAFLD were obtained from the GEO database. And the disease genes for NAFLD and intestinal flora disorder were obtained from the disease databases. The protein-protein interaction network was established by string 11.0 database and visualized by Cytoscape 3.7.2 software. Cytoscape plug-in MCODE and cytoHubba were used to screen the potential genes of intestinal flora disorder and NAFLD, to obtain potential targets for intestinal flora to interfere in the occurrence and process of NAFLD. Enrichment analysis of potential targets was carried out using R 4.0.2 software.
Results:
The results showed that 7 targets might be the key genes for intestinal flora to interfere with NAFLD. CCL2, IL6, IL1B, and FOS are mainly related to the occurrence and development mechanism of NAFLD, while PTGS2, SPINK1, and C5AR1 are mainly related to the intervention of intestinal flora in the occurrence and development of NAFLD. The gene function is mainly reflected in basic biological processes, including the regulation of metabolic process, epithelial development, and immune influence. The pathway is mainly related to signal transduction, immune regulation, and physiological metabolism. The TNF signaling pathway, AGE-RAGE signaling pathway in diabetic activity, and NF-Kappa B signaling pathways are important pathways for intestinal flora to interfere with NAFLD. According to the analysis results, there is a certain correlation between intestinal flora disorder and NAFLD.
Conclusion:
It is speculated that the mechanism by which intestinal flora may interfere with the occurrence and development of NAFLD is mainly related to inflammatory response and insulin resistance. Nevertheless, further research is needed to explore the specific molecular mechanisms.
Keywords: bioinformatics, intestinal flora disorder, nonalcoholic fatty liver disease, signaling pathway, target prediction
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is becoming increasingly prevalent and rapidly becoming the leading cause of chronic liver disease worldwide. Around the world, 25.2% of the population suffers from NAFLD, and with the increase in the prevalence rate, NAFLD remains the most common liver disease in the world.[1,2] NAFLD mainly includes simple steatosis (SS), non-alcoholic steatohepatitis (NASH), and its associated cirrhosis (NASH cirrhosis). NAFLD is characterized by liver fat accumulation of more than 5% in the absence of any secondary cause.[2,3] NASH is a kind of liver cell damage caused by necrotizing inflammation under the premise of liver steatosis, which can easily develop into cirrhosis and liver cancer. NAFLD is a major cause of chronic liver disease. Under the influence of diet, lifestyle, and other factors, the incidence rate increases year by year, and the probability of liver cell progression from simple steatosis to hepatitis, cirrhosis, and hepatocellular carcinoma increases.[4,5]
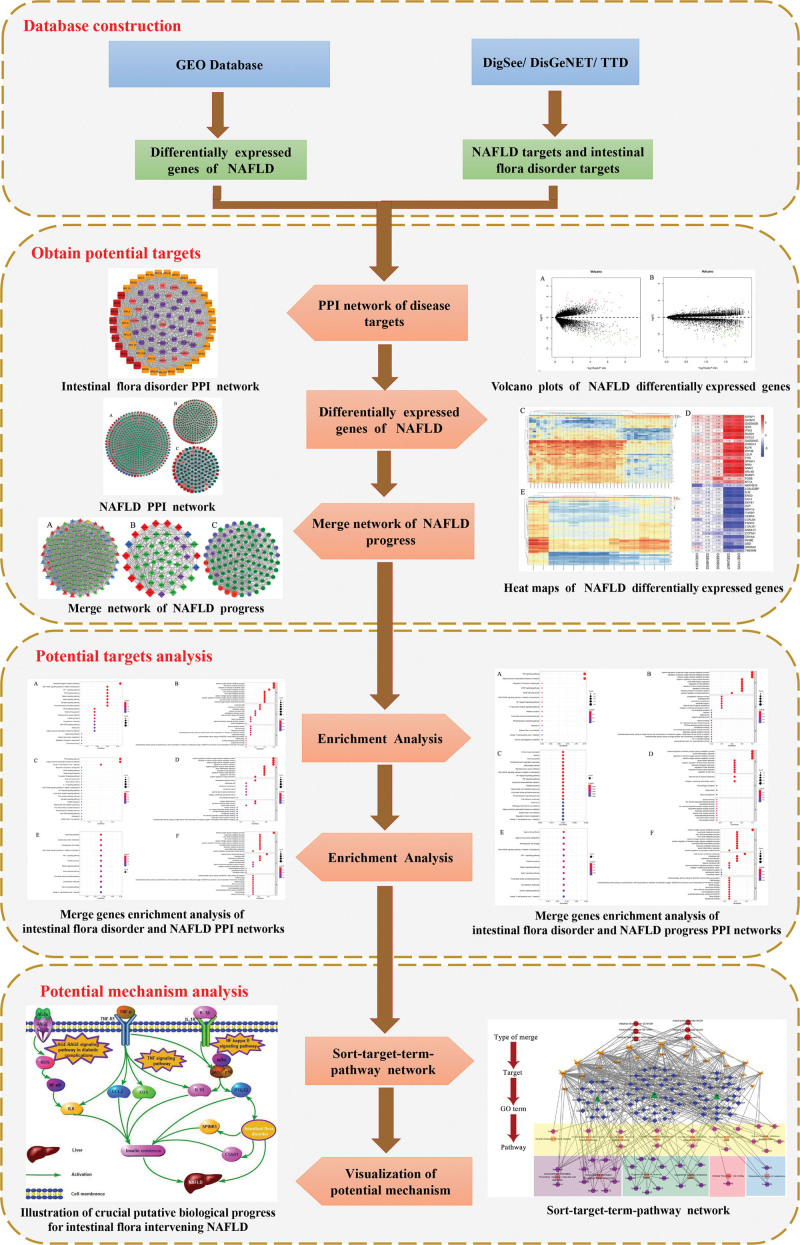
Intestinal flora is the general name of the intestinal microbial community, and the total number of genes encoded by this complex microbial community is 150 times that of the number of genes encoded by human.[6,7] The function of intestinal flora is not only related to nutrition digestion but also closely related to immunity and metabolic diseases with the in-depth study.[8,9] In 1998, Marshall first proposed the theory of “gut–liver axis”,[10] that is, after the intestinal barrier is damaged, bacterial translocation endotoxin enters the portal vein system, activates liver Kupffer cells, etc., and then releases a series of inflammatory factors, further aggravating liver damage and disease progression. Many studies[11,12] have shown that intestinal flora disorder is associated with NAFLD. The composition of intestinal flora varies from SS to NASH, fibrosis, and cirrhosis. Therefore, intestinal flora may be a predictor of NAFLD progression and severity. The involvement of microorganisms in changing the balance between pro-inflammatory or anti-inflammatory signals may lead to the progression of NASH inflammation.[13,14] The pathogenesis of NAFLD remains unclear, and it is thought to involve a complex interaction between genetic susceptibility variation, environmental factors, insulin resistance, and changes in intestinal flora.[15] There is an urgent need to understand the relationship between NAFLD and intestinal flora disorder, which may serve as predicators of NAFLD progression and severity, and may contribute to improved diagnosis, patient stratification, and identification of new therapeutic targets. To this end, we investigated the relationship between intestinal flora disorder and non-alcoholic fatty liver disease based on bioinformatics. This paper mainly explores the relationship between intestinal flora disorder and SS, NASH, and NASH cirrhosis. The specific workflow is displayed in Figure 1.
Figure 1.
Workflow of bioinformatics.
2. Materials and Methods
2.1. Collect disease targets associated with intestinal flora disorder
Intestinal flora disorder, intestinal dysbiosis, intestinal flora imbalance, intestinal microbiota disorder, and dysbiosis of intestinal flora are taken as retrieval words. Proteins related to intestinal flora disorder are collected through the disease databases. Including Therapeutic Targets Database (TTD) (https://db.idrblab.org/ttd/),[16] DigSee (http://210.107.182.61/geneSearch/)[17] and DisGeNET (http://www.disgenet.org/search).[18] Species were selected as “Homo sapiens”, and gene-disease score was set ≥ 0.1 in DisGeNET database. 80 proteins related to intestinal flora disorder were finally obtained.
2.2. Collect disease targets associated with NAFLD
Screening of differentially expressed genes (DEGs) in NAFLD: search in GEO database (https://www.ncbi.nlm).[19] And use the following keywords to screen gene expression profile: non-alcoholic fatty liver disease; the tissue source is “human liver tissue”; the research type is “Expression profiling by Array”; nonalcoholic fatty liver in the experimental group and healthy liver in the control group. Finally, we have acquired one SS gene chip (GSE89632), five NASH gene chips (GSE17470, GSE24807, GSE33814, GSE89632, GSE48452), and one NASH cirrhosis gene chip (GSE58979). Among the GSE58979 microarray samples, it is considered to be a normal control group for steatosis of less than 5%. And screen NASH cirrhosis DEGs from visceral samples. R 4.0.2 software was used to screen DEGs. During the screening process, the screening condition was log2 (FC) > 1.5, P ≤ .05. Finally, we obtained 93 DEGs of SS, 51 DEGs of NASH, and 53 DEGs of NASH cirrhosis.
Collection of disease proteins associated with NAFLD: NAFL, simple fatty liver, steatosis, NASH, steatohepatitis, NASH cirrhosis, and NASH-cirrhosis were searched in the disease databases for retrieval words. We collect disease targets related to SS, NASH, and NASH cirrhosis in the Therapeutic Target Database (TTD) (https://db.idrblab.org/ttd/), DigSee (http://210.107.182.61/geneSearch/) and DisGeNET (http://www.disgenet.org/search). Species were selected as “Homo sapiens”, and gene-disease score was set ≥ 0.1 in DisGeNET database. After removing repetitive disease targets, 643 SS-related proteins, 333 NASH-related proteins, and 120 NASH cirrhosis-related proteins can be acquired in the end. Combining all NAFLD genes, we finally gained 718 SS-related targets, 379 NASH-related targets, and 171 NASH cirrhosis relates targets.
2.3. Construction of protein-protein interaction network and screening of key genes
String 11.0 database (https://string- db.org/) is a database that stores known and predicted protein interactions, including direct and indirect interactions of proteins. It scores the information of each protein interaction. The higher the score, the higher the confidence of protein interaction.[20] Protein names related to intestinal flora disorder and NAFLD collected in the disease database were input into the String 11.0 database for retrieval. And protein interaction data with a high confidence interval score greater than 0.7 were selected to ensure the reliability of the data. The obtained protein interaction data were imported into Cytoscape 3.7.2 (http://www.cytoscape.org/) software to construct protein–protein interaction (PPI) network related to intestinal flora disorder and NAFLD. Cytoscape is an open-source bioinformatics analysis software used to construct molecular interaction networks composed of protein, gene, and drug interactions for visual browsing and analysis.[21]
The key targets of disease are screened using MCODE plug-ins and cytoHubba plug-ins included in the Cytoscape 3.7.2 software. The MCODE plug-in was used for module analysis to select the genes of the first two modules with high scores. Screen Hub genes using cytoHubba plug-in. The genes screened by the above two methods were intersected to obtain the key targets for the treatment of diseases.
By using the Merge function of Cytoscape 3.7.2 software, the PPI network of intestinal flora disorder is merged with SS, NASH, and NASH cirrhosis PPI network respectively. To select the intersecting parts of the network, proteins that exist in the intersection may be a potential core target for intestinal flora to interfere with NAFLD. These potential core targets were systematically analyzed to explore the relationship between intestinal flora and NAFLD. The PPI network of NAFLD was separately merged to find the connection between different stages of NAFLD diseases at the intersection, to help explore the relationship between intestinal flora and NAFLD.
2.4. Gene ontology (and Kyoto encyclopedia of genes and genomes pathway enrichment
To evaluate the role of potential core targets by bioinformatic annotation, the R 4.0.2 (https://cran.r-project.org/doc/FAQ/R-FAQ.html#Citing-R) software with the Bioconductor package was manipulated, including gene ontology (GO) knowledgebase (http://geneontology.org/), Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis (https://www.genome.jp/kegg/).[22–24]
3. Results
3.1. Intestinal flora disorder PPI network
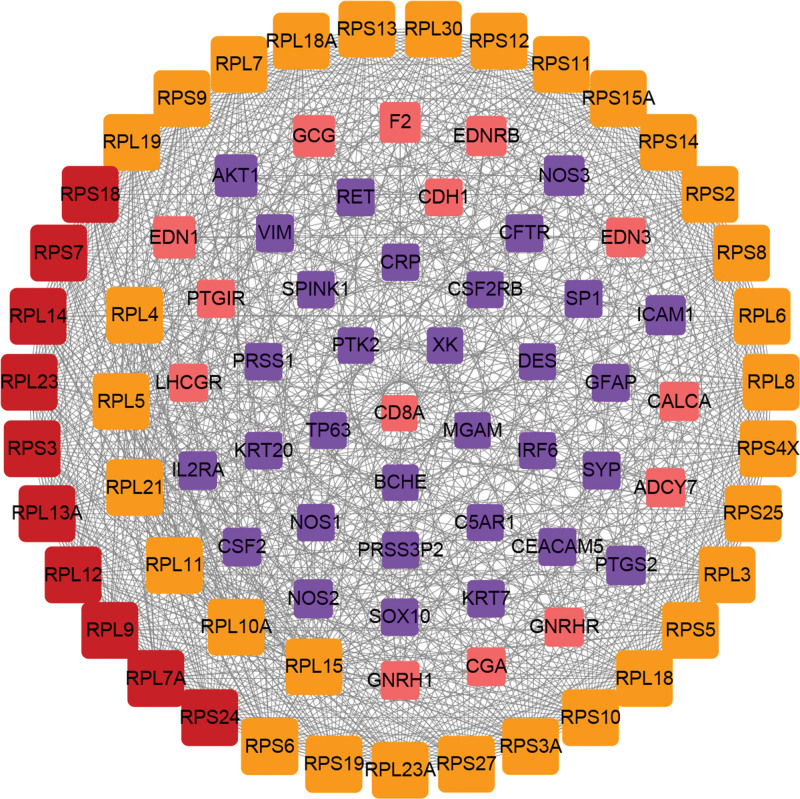
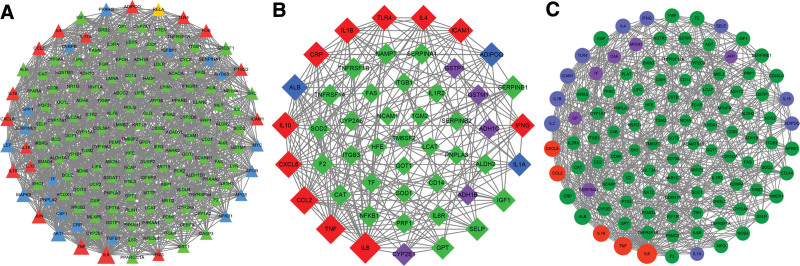
Introduce 80 disease targets related to intestinal flora disorder obtained from the database into String 11.0 database, and set high confidence >0.7. In addition, “the 1st shell” and “the 2nd shell” were set to “no more than 20 interactors” in this study. The protein interaction data were then visualized by Cytoscape 3.7.2 software to obtain the PPI network of intestinal flora disorder (Fig. 2). The network had 86 nodes, which interacted with 930 edges. The size of nodes is positively correlated with the degree value of nodes. Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (41 genes) and module 2 (12 genes) and intersect the first 10 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to intestinal flora disorder. According to the above method, a total of 10 key targets related to intestinal flora disorder were produced (as shown in Table 1, Supplemental Digital Content, http://links.lww.com/MD/H65).
Figure 2.
Intestinal flora disorder PPI network. Yellow nodes represent module 1 genes, pink nodes represent module 2 genes, blue nodes represent hub genes, and red nodes represent intersection genes of modules and hub. PPI = protein–protein interaction.
3.2. DEGs screening of NAFLD
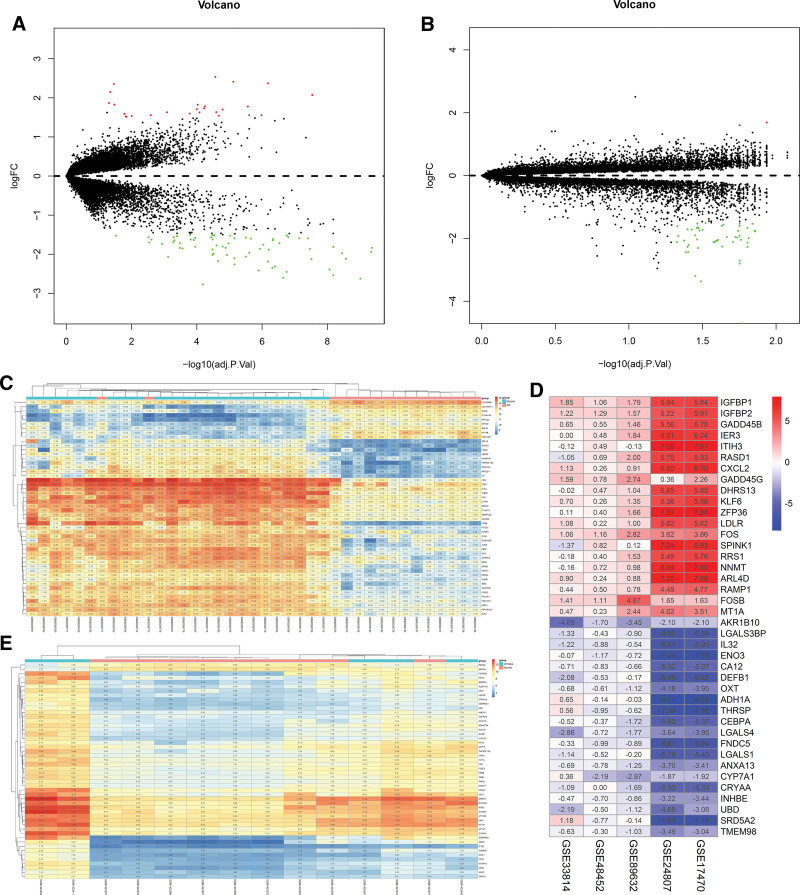
The gene chips obtained from the GEO database were analyzed by R 4.0.2 (Fig. 3). In the volcano plots, the red nodes represent upregulated genes and green nodes represent downregulated genes. In the heat maps, red areas represent upregulated genes and blue or green represents downregulated genes. According to the adjusted criteria of P≤0.05 and |log2 (FC) |≥1.5, 93 DEGs were selected from GSE89632 chip (67 down-regulated genes, 26 up-regulated genes). 50 integrated DEGs were screened from GSE17470, GSE24807, GSE33814, GSE89632, and GSE48452 (24 down-regulated genes, 26 upregulated genes). And 53 DEGs were screened out in GSE58979 (51 down-regulated genes, 2 upregulated genes). Information on differentially expressed genes is shown in Table 2, Supplemental Digital Content, http://links.lww.com/MD/H65.
Figure 3.
Volcano plot and Heat map of NAFLD differentially expressed genes. (A) Volcano plot of SS; (B) Volcano plot of NASH cirrhosis; (C) Heat map of SS; (D) Heat map of NASH; (E) Heat map of NASH cirrhosis. NAFLD = nonalcoholic fatty liver disease, NASH = nonalcoholic steatohepatitis.
3.3. NAFLD PPI network
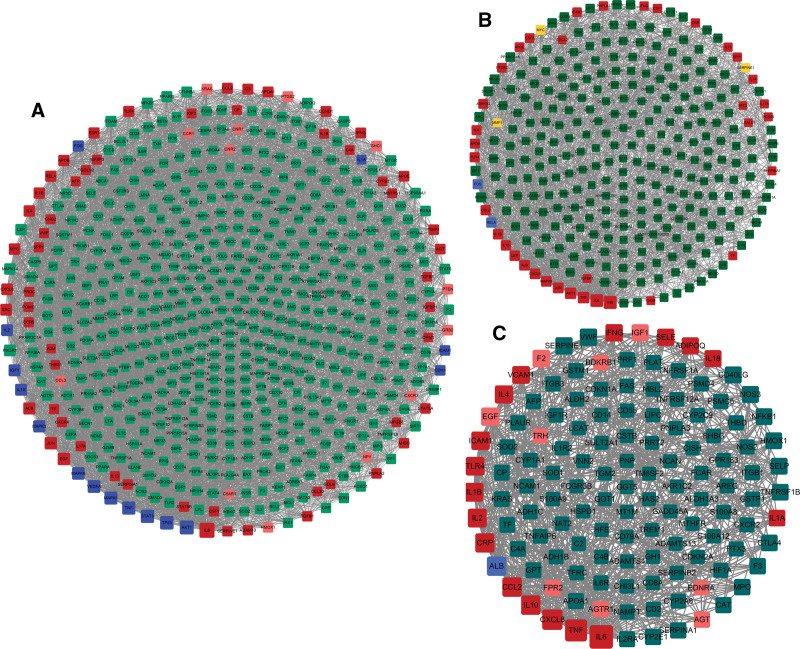
718 SS disease targets, 379 NASH-related targets, and 171 NASH cirrhosis relates targets were input into the String 11.0 database, respectively, with high confidence greater than 0.7. And the obtained protein interaction data were imported into Cytoscape 3.7.2 software to build SS PPI network (Fig. 4A), NASH PPI network (Fig. 4B) and NASH cirrhosis PPI network (Fig. 4C). The module analysis of the MCODE plugin and the hub genes of cytoHubba plugin were used to screen the key disease targets of these three PPI networks. The key targets of the disease are obtained by intersecting the genes of the first two modules and the top 10% of the hub genes.
Figure 4.
NAFLD PPI network. (A) SS PPI network; (B) NASH PPI network; (C) NASH cirrhosis PPI network. Yellow nodes represent the genes of module 1, pink nodes represent the genes of module 2, blue nodes represent hub genes, and red nodes represent the intersection of module genes and hub genes. The size of nodes is positively correlated with the degree value of nodes. NAFLD = nonalcoholic fatty liver disease, NASH = nonalcoholic steatohepatitis, PPI = protein–protein interaction.
The PPI network of SS has 591 nodes and 5295 edges. Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (25 genes) and module 2 (45 genes) and intersect the first 72 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to SS. According to the above method, a total of 57 key targets related to SS were produced (as shown in Table 1, Supplemental Digital Content, http://links.lww.com/MD/H65). The PPI network of NASH has 300 nodes and 1746 edges. Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (18 genes) and module 2 (21 genes) and intersect the first 38 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to NASH. According to the above method, a total of 36 key targets related to NASH were produced (as shown in Table 1, Supplemental Digital Content, http://links.lww.com/MD/H65). The PPI network of NASH cirrhosis has 128 nodes and 630 edges. Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (17 genes) and module 2 (9 genes) and intersect the first 18 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to NASH. According to the above method, a total of 17 key targets related to NASH were produced (as shown in Table 1, Supplemental Digital Content, http://links.lww.com/MD/H65).
3.4. Merge of intestinal flora disorder PPI network and NAFLD PPI network
The PPI network of intestinal flora disorder and the PPI network of SS, NASH, and NASH cirrhosis were merged through the merge function of Cytoscape 3.7.2 software, respectively. And the possible targets of intestinal flora in the treatment of NAFLD were found. We have obtained 20 possible targets for treating SS, 7 for treating NASH, and 7 for treating NASH cirrhosis. These are potential targets for intestinal flora to intervene in different stages of NAFLD. The information of merge genes is shown in Table 1.
Table 1.
Information of merge networks genes.
| Merge target | Official Full Name | Type of merge |
|---|---|---|
| CCL2 | C-C motif chemokine ligand 2 | SS-NASH、NASH-NASH cirrhosis、SS-NASH cirrhosis |
| TNF | Tumor necrosis factor | SS-NASH、NASH-NASH cirrhosis、SS-NASH cirrhosis |
| TLR4 | Toll-like receptor 4 | SS-NASH、NASH-NASH cirrhosis |
| CXCL8 | C-X-C motif chemokine ligand 8 | SS-NASH、NASH-NASH cirrhosis、SS-NASH cirrhosis |
| IL4 | Interleukin 4 | SS-NASH、NASH-NASH cirrhosis |
| IL1A | Interleukin 1 alpha | SS-NASH |
| ICAM1 | Intercellular adhesion molecule 1 | SS-NASH、NASH-NASH cirrhosis、intestinal flora disorder -SS、intestinal flora disorder -NASH、intestinal flora disorder -NASH cirrhosis |
| IFNG | Interferon gamma | SS-NASH、NASH-NASH cirrhosis |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | SS-NASH、intestinal flora disorder -SS、intestinal flora disorder -NASH |
| IL13 | Interleukin 13 | SS-NASH |
| IL6 | Interleukin 6 | SS-NASH、NASH-NASH cirrhosis、SS-NASH cirrhsosis |
| IL1B | Interleukin 1 beta | SS-NASH、NASH-NASH cirrhosis |
| IL17A | Interleukin 17A | SS-NASH |
| IL10 | Interleukin 10 | SS-NASH、NASH-NASH cirrhosis、SS-NASH cirrhsosis |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit | SS-NASH |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit | SS-NASH |
| ADIPOQ | Adiponectin, C1Q and collagen domain containing | SS-NASH |
| CRP | C-reactive protein | NASH-NASH cirrhosis、intestinal flora disorder -SS、intestinal flora disorder -NASH、intestinal flora disorder -NASH cirrhosis |
| EDN1 | Endothelin 1 | intestinal flora disorder -SS |
| AKT1 | AKT serine/threonine kinase 1 | intestinal flora disorder -SS、intestinal flora disorder -NASH |
| NOS3 | Nitric oxide synthase 3 | intestinal flora disorder -SS、intestinal flora disorder -NASH cirrhosis |
| CSF2RB | Colony stimulating factor 2 receptor subunit beta | intestinal flora disorder -SS |
| IL2RA | Interleukin 2 receptor subunit alpha | intestinal flora disorder -SS、intestinal flora disorder -NASH cirrhosis |
| CALCA | Calcitonin related polypeptide alpha | intestinal flora disorder -SS |
| F2 | Coagulation factor II, thrombin | intestinal flora disorder -SS、intestinal flora disorder -NASH、intestinal flora disorder -NASH cirrhosis |
| CD8A | CD8a molecule | intestinal flora disorder -SS、intestinal flora disorder -NASH cirrhosis |
| KRT20 | Keratin 20 | intestinal flora disorder -SS |
| CDH1 | Cadherin 1 | intestinal flora disorder -SS |
| PRSS1 | Serine protease 1 | intestinal flora disorder -SS |
| SPINK1 | Serine peptidase inhibitor Kazal type 1 | intestinal flora disorder -SS |
| NOS2 | Nitric oxide synthase 2 | intestinal flora disorder -SS、intestinal flora disorder -NASH cirrhosis |
| IRF6 | Interferon regulatory factor 6 | intestinal flora disorder -SS |
| SP1 | Sp1 transcription factor | intestinal flora disorder -SS |
| C5AR1 | Complement C5a receptor 1 | intestinal flora disorder -SS |
| BCHE | Butyrylcholinesterase | intestinal flora disorder -SS |
| CSF2 | Colony stimulating factor 2 | intestinal flora disorder -NASH |
| GCG | Glucagon | intestinal flora disorder -NASH |
3.5. Merge network of NAFLD progress
The SS PPI network, NASH PPI network, and NASH cirrhosis PPI network are merged by the merge function of Cytoscape software respectively, to find key targets for NAFLD development. The module analysis of the MCODE plugin and the hub genes of cytoHubba plugin was used to screen the key progress targets of NAFLD. The key progress targets of NAFLD are obtained by intersecting the first two modules and the top 10% of the hub genes, in the merge network.
The merge network of SS PPI network and NASH PPI network has 171 nodes and 1044 edges (as shown in Fig. 5A). Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (27 genes) and module 2 (9 genes) and intersect the first 18 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to the progression from SS to NASH. Based on the above approach, a total of 17 key targets related to the progression from SS to NASH were obtained. The merge network of NASH PPI network and NASH cirrhosis PPI network has 50 nodes and 207 edges. (as shown in Fig. 5B) Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (14 genes) and module 2 (5 genes) and intersect the first 11 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to the progression from NASH to NASH cirrhosis. Based on the above approach, a total of 11 key targets related to the progression from NASH to NASH cirrhosis were obtained. The merge network of SS PPI network and NASH cirrhosis PPI network has 108 nodes and 530 edges. (as shown in Fig. 5C) Module analysis was conducted through the MCODE plugin. Select the genes for module 1 (15 genes) and module 2 (6 genes) and intersect the first 5 hub genes obtained from cytoHubba plugin to obtain the key disease targets related to the progression from SS to NASH cirrhosis. Based on the above approach, a total of 5 key targets related to the progression from SS to NASH cirrhosis were obtained. The information of merge genes is shown in Table 1.
Figure 5.
Merge network of NAFLD progress. (A) Merge network of SS and NASH PPI networks; (B) Merge network of NASH and NASH cirrhosis PPI networks; (C) Merge network of SS and NASH cirrhosis PPI networks. Blue nodes represent module 1 genes, purple nodes represent module 2 genes, yellow nodes represent hub genes, and red nodes represent the intersection of module genes and hub genes. The size of nodes is positively correlated with the degree value of nodes. NAFLD = nonalcoholic fatty liver disease, NASH = nonalcoholic steatohepatitis, PPI = protein–protein interaction.
The intestinal flora disorder PPI network was merged with the merge network of NAFLD progress through the Merge function of Cytoscape 3.7.2 software, and the potential targets for intestinal flora interfering NAFLD progress were found. We obtained 5 potential targets (AKT1, F2, ICAM1, PTGS2, CRP) for intestinal flora to intervene the progression of NAFLD from SS to NASH, 3 potential targets (CRP, ICAM1, F2) for intestinal flora to intervene the progression of NAFLD from NASH to NASH cirrhosis, and 7 potential targets (NOS3, IL2RA, F2, CD8A, NOS2, ICAM1, CRP) for intestinal flora to intervene the progression of NAFLD from SS to NASH cirrhosis were obtained. These are considered as potential targets for intestinal flora to intervene in the NAFLD process.
Subsequently, we compared all merge genes with DEGs of NAFLD and found 7 overlapping targets (CCL2, PTGS2, IL6, IL1B, FOS, SPINK1, C5AR1). In this study, these 7 targets were the core potential targets for intestinal flora to intervene in NAFLD.
3.6. GO function enrichment and KEGG pathway enrichment analysis
We used R 4.0.2 (https://cran.r-project.org/doc/FAQ/R-FAQ.html#Citing-R) software to perform GO and KEGG enrichment analysis on the protein targets of the merge networks. In the bubble chart, the X-axis represents the number of target genes (Gene Ratio), and the Y-axis represents the KEGG pathway or GO term where the target gene is significantly enriched. The size of the dots intuitively reflects the size of the Gene Ratio, and the color depth of the dots reflects different P value ranges.
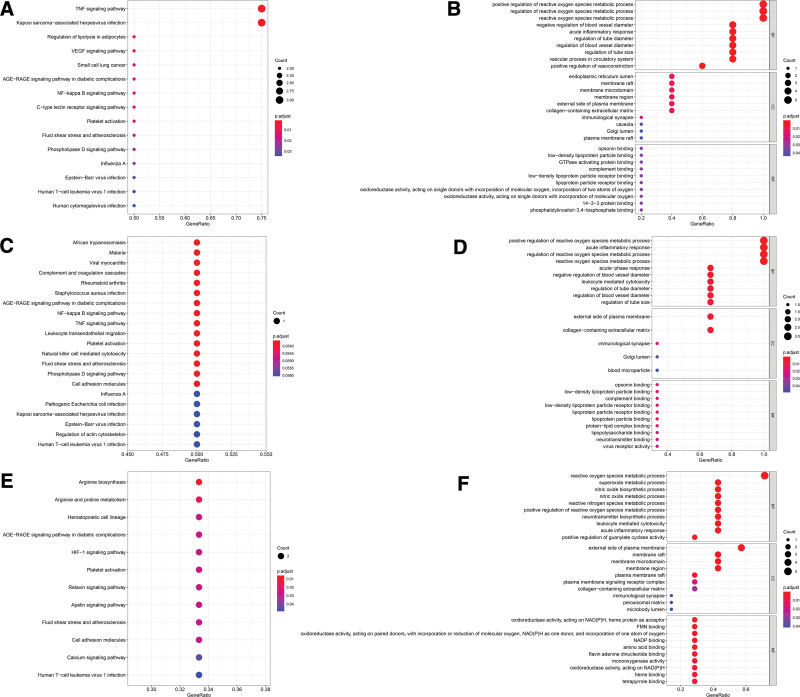
The merge genes enrichment analysis of intestinal flora disorder and NAFLD PPI network is as shown in Figure 6. Analysis of merge of intestinal flora disorder and SS PPI networks: (1) KEGG pathway enrichment analysis (Fig. 6A) found 148 pathways, of which 20 pathways had P value and q value less than .05. In the GO enrichment analysis (Fig. 6B), a total of 541 terms were found, of which 512 terms were related to biological processes (BP), 11 terms were related to cell composition (CC) and 18 terms were related to molecular function (MF); analysis of merge of intestinal flora disorder and NASH PPI networks: KEGG pathway enrichment analysis (Fig. 6C) found 121 pathways, of which 22 pathways had P value and q value less than .05. In the GO enrichment analysis (Fig. 6D), a total of 616 terms were found, of which 590 terms were related to biological processes (BP), 7 terms were related to cell composition (CC) and 19 terms were related to molecular function (MF). (3) Analysis of merge of intestinal flora disorder and NASH cirrhosis PPI networks: KEGG pathway enrichment analysis (Fig. 6E) found 57 pathways, of which 12 pathways had P value and q value less than .05. In the GO enrichment analysis (Fig. 6F), a total of 347 terms were found, of which 305 terms were related to biological processes (BP), 10 terms were related to cell composition (CC) and 32 terms were related to molecular function (MF).
Figure 6.
Merge genes enrichment analysis of intestinal flora disorder and NAFLD PPI networks. (A) KEGG pathway analysis for merge of intestinal flora disorder and SS PPI networks; (B) GO enrichment analysis for merge of intestinal flora disorder and SS PPI networks; (C) KEGG pathway analysis for merge of intestinal flora disorder and NASH PPI networks; (D) GO enrichment analysis for merge of intestinal flora disorder and NASH PPI networks; (E) KEGG pathway analysis for merge of intestinal flora disorder and NASH cirrhosis PPI networks; (F) GO enrichment analysis for merge of intestinal flora disorder and NASH cirrhosis PPI networks. GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes.
The merge genes enrichment analysis of merge network of NAFLD progress is as shown in Figure 7. Analysis of merge of intestinal flora disorder, SS, and NASH PPI networks: (1) KEGG pathway enrichment analysis (Fig. 7A) found 115 pathways, of which 15 pathways had P value and q value less than .05. In the GO enrichment analysis (Fig. 7B), a total of 670 terms were found, of which 636 terms were related to biological processes (BP), 12 terms were related to cell composition (CC) and 22 terms were related to molecular function (MF); analysis of merge of intestinal flora disorder, NASH and NASH cirrhosis PPI networks: KEGG pathway enrichment analysis (Fig. 7C) found 22 pathways, of which 21 pathways P value and q value less than .06. In the GO enrichment analysis (Fig. 7D), a total of 323 terms were found, of which 299 terms were related to biological processes (BP), 5 terms were related to cell composition (CC) and 19 terms were related to molecular function (MF); analysis of merge of intestinal flora disorder, SS and NASH cirrhosis PPI networks: KEGG pathway enrichment analysis (Fig. 7E) found 57 pathways, of which 12 pathways had P value and q value less than .05. In the GO enrichment analysis (Fig. 7F), a total of 347 terms were found, of which 305 terms were related to biological processes (BP), 10 terms were related to cell composition (CC) and 32 terms were related to molecular function (MF).
Figure 7.
Merge genes enrichment analysis of intestinal flora disorder and NAFLD progress PPI networks. (A) KEGG pathway analysis for merge of intestinal flora disorder, SS and NASH PPI networks; (B) GO enrichment analysis for merge of intestinal flora disorder, SS and NASH PPI networks; (C) KEGG pathway analysis for merge of intestinal flora disorder, NASH and NASH cirrhosis PPI networks; (D) GO enrichment analysis for merge of intestinal flora disorder, NASH and NASH cirrhosis PPI networks; (E) KEGG pathway analysis for merge of intestinal flora disorder, SS and NASH cirrhosis PPI networks; (F) GO enrichment analysis for merge of intestinal flora disorder, SS and NASH cirrhosis PPI networks. GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes.
GO functional enrichment analysis showed that the gene functions of intestinal flora interfering with NAFLD are mainly reflected in BP, including regulating metabolic processes, epithelial development, and affecting immunity. KEGG enrichment analysis found that the pathway intervention of intestinal flora in NAFLD was mainly closely related to signal transduction, immune regulation, and physiological metabolism.
3.7. Sort-target-term-pathway network
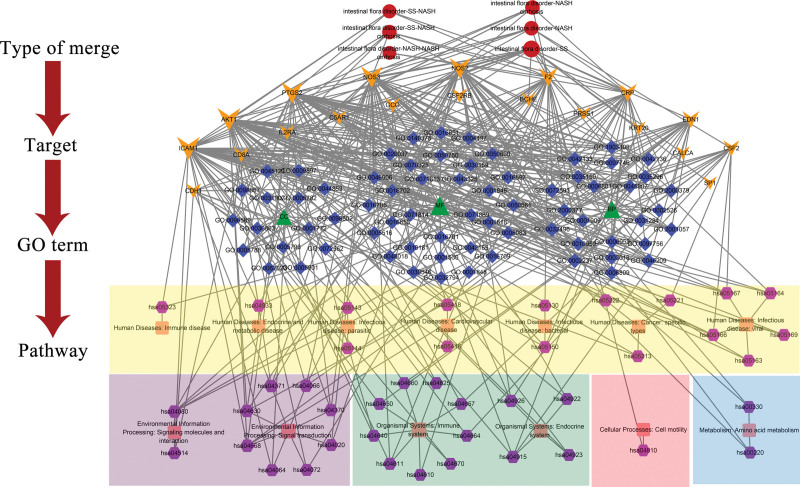
Figure 8 was obtained by visualizing the merge genes, GO terms, and KEGG pathways through Cytoscape 3.7.2 software. The network has 153 nodes (6 nodes for merge sort, 20 nodes for merge genes, 3 nodes for GO sort, 69 nodes for GO terms, 13 nodes for KEGG BRITE, 42 nodes for KEGG pathways) and 480 edges. The targets with degree value greater than 10 were AKT1, ICAM1, NOS3, PTGS2, NOS2, F2, CRP, EDN1, CSF2, CD8A, and CDH1. The top 10 GO terms of degree value are mainly BP terms (response to molecule of bacterial origin, response to lipopolysaccharide, reactive oxygen species metabolic process, neurotransmitter metabolic process, nitric oxide biosynthetic process, neurotransmitter biosynthetic process, nitric oxide metabolic process, reactive oxygen species biosynthetic process, positive regulation of reactive oxygen species metabolic process, and reactive nitrogen species metabolic process). The KEGG pathway with a higher degree value is mainly related to signal transduction (TNF signaling pathway, HIF-1 signaling pathway, Apelin signaling pathway, JAK-STAT signaling pathway), human disease (AGE-RAGE signaling pathway in diabetic complications, Human T-cell leukemia virus 1 infection, Kaposi sarcoma-associated herpesvirus infection, Fluid shear stress, and atherosclerosis) and endocrine system (Estrogen signaling pathway, Relaxin signaling pathway).
Figure 8.
Sort-target-term-pathway network. Red round nodes represent type of merge. Yellow inverted triangle nodes represent merge genes. Green triangle nodes represent type of GO terms. Blue diamond nodes represent GO terms. Pink square nodes represent the BRITE of KEGG. Purple hexagon nodes represent KEGG pathways. The size of nodes is positively correlated with the degree value of nodes. GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes.
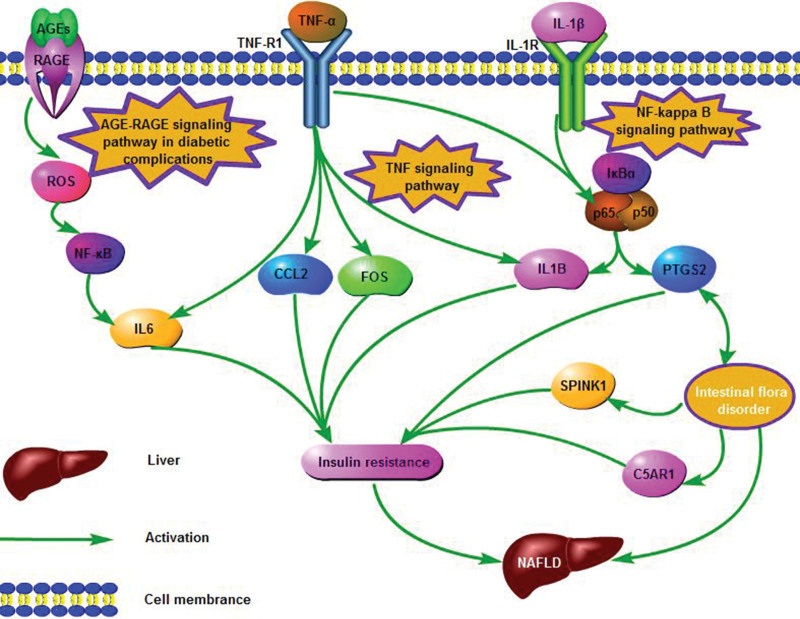
Interference of intestinal flora with the pathological process of NAFLD is closely associated with inflammation and insulin resistance. TNF signaling pathway, AGE-RAGE signaling pathway in the diabetic activity, and NF-kappa B signaling pathway will promote the up-regulation of CCL2, IL6, IL1B, FOS, SPINK1, C5AR1, and PTGS2 after activation, which will lead to liver inflammation and promote the occurrence and development of NAFLD. Intestinal flora can act on SPINK1, C5AR1, and PTGS2 to improve NAFLD. CCL2, IL6, IL1B, FOS, and NF-κB may play an important role in the occurrence and development of NAFLD. KEGG Database (https://www.kegg.jp/kegg/kegg1.html) and software of Pathway Builder Tool 2.0 were used to generate the figure. As seen in Figure 9, the major predictive signaling pathways for intestinal flora interfering with NAFLD were constructed.
Figure 9.
Illustration of crucial putative biological progress for intestinal flora intervening NAFLD. NAFLD = nonalcoholic fatty liver disease.
4. Discussion
NAFLD is a manifestation of obesity and metabolic syndrome affecting the liver, and its pathogenesis is associated with numerous factors.[2] The exact pathogenesis of NAFLD has not been fully elucidated. The current theories that are mainly concerned are the second hit theory and the gut-liver axis theory. Intestinal flora has a strong effect on the liver, and can participate in the development and progression of NAFLD through the “gut-liver axis”.[25–27] Most scholars believe that factors related to the pathogenesis of NAFLD include immune activation,[28] inflammatory response,[29] bile acid metabolism interference,[30] insulin resistance,[31] and fasting-induced adipose factor.[32]
CCL2, also known as monocyte chemoattractant protein 1, is mainly secreted by monocytes and macrophages.[33] Mouse experiments[34,35] showed that when the chemokine CCL2 was overexpressed, macrophages in adipose tissue were recruited to secrete a large number of inflammatory factors, promoting liver steatosis, inflammation, and fibrosis. CCL2 deficiency can resist lipid accumulation and insulin resistance induced by a high-fat diet. Many studies[36,37] have suggested that CCL2 expression is upregulated in the liver of animal models of NAFLD, and its circulating level in NAFLD patients is also increased. NAFLD can gradually develop from SS and NASH to NASH cirrhosis and even liver cancer. CCL2 can affect the development and progression of NAFLD by participating in processes such as steatosis and inflammation. Il-6, which is produced by T cells, B cells, macrophages, and endothelial cells, is a cytokine with a variety of physiological effects and an important inflammatory factor, induces the production of TNF-α, and plays an autocrine role in the production of other pro-inflammatory cytokines.[38] Clinically relevant studies have reported significantly elevated serum IL-6 and TNF-α levels in NAFLD patients, suggesting that high levels of TNF-α and IL-6 may be involved in the occurrence and development of NAFLD.[39] A large number of foreign studies have also shown that there are significant immune disorders in the surrounding tissues and liver of NAFLD patients, accompanied by excessive production of inflammatory factors such as TNF-α and IL-6.[40,41] In-depth studies have shown that IL-6 can lead to liver steatosis, insulin resistance, and aggravation of inflammation.[39,42] Peripheral adipocytes and hepatocytes can secrete IL-6, which in turn mediates and promotes macrophage infiltration, thereby participating in the development and progression of NAFLD.[39] The protein encoded by IL1B is a member of the interleukin 1 cytokine family. This cytokine is an important mediator of the inflammatory response and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis.[43,44] Liver Kupffer cells can produce IL-1B, which exacerbates liver inflammation and steatosis.[45] IL-1B promotes the production of nitric oxide and cell apoptosis in islet cells, leading to selective destruction of islet cells, thus further inducing insulin resistance.[45] In addition to playing a role in inflammation, IL1B may contribute to NAFLD pathogenesis by promoting insulin resistance and altering lipid metabolism.[46,47] FOS is considered a regulator of cell proliferation, differentiation, and transformation, which is closely related to inflammatory response and tumor. FOS can encode leucine zipper proteins that can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1.[48] Christoph’s work revealed significant changes in genes associated with metabolic processes, transport, signal transduction, and redox in the mouse NASH model, and AP-1 appears to be the key transcriptional regulator of these changes.[49] FOS expression is related to the occurrence of insulin resistance.[50] Significantly increased DNA binding of NF-κB and AP-1 in NASH patients plays a major role in oxidative stress and insulin resistance of NASH pathophysiological mechanisms.[51] SPINK1, as a secretory trypsin inhibitor, is a potential marker for the diagnosis of liver cancer.[52] SPINK1 is expressed in both the pancreas and the gastrointestinal tract, and the trypsin inhibitor encoded by this gene can be secreted from pancreatic acinar cells into pancreatic juice, which is often present at high levels in the pancreas and pancreatic juice.[53–55] In the normal gastrointestinal tract, SPINK1 is thought to play a protective role in both gastric mucosa[56] and colonic mucosa.[57] Evidence shows that SPINK1 is an important growth factor linking chronic inflammation and cancer,[58–60] and the progression of NAFLD to liver cancer and cell cancer may be driven by SPINK-1 gene.[55,61,62] Complement receptors CR1 and CR3 are responsible for the phagocytic and adhesive properties of neutrophils, whereas the C5a receptor mediates the pro-inflammatory and chemotactic actions of the C5aR1.[63] C5aR1 has been shown to promote primary tumor growth and immunosuppression.[64,65] C5aR1-mediated tumor promotion and immunosuppression have been attributed to the C5aR1 ligand C5a.[66,67] Studies have shown that reducing C5 levels can reduce inflammatory responses, thus significantly reducing the degree of liver fibrosis and cirrhosis.[68,69] Sendler et al.[70] used C5a receptor antagonist to inhibit C5aR1 and found that it could greatly reduce fibrogenesis after pancreatic necrosis. PTGS2 is the principal isozyme responsible for the production of inflammatory prostaglandins and plays an important role in inflammation and proliferation of a variety of cells and tissues. PTGS2 and its products affect the metabolism of fat, carbohydrate, and protein, thus interfering with the metabolism of normal substances in the liver and affecting the occurrence and development of liver disease.[71] The increased expression of PTGS2 causes triglycerides to accumulate in liver cells,[72] which may lead to insulin resistance. Insulin resistance can lead to increased free fatty acids and transport to liver cells for accumulation, increased release of inflammatory factors, and up-regulated expression of PTGS2.[73] Inhibition of PTGS2 expression may be related to the prevention of liver histological changes caused by intestine endotoxin.[74,75]
In addition, studies have shown that the intestinal microbiome-driven PTGS2 pathway plays a key role in suppressing tumor immunity, suggesting that PTGS2 and its receptor may be new targets for the treatment of NAFLD.[76] C5AR1 can induce the release of various immune inflammatory mediators and cytokines, which help to recruit and activate immune and inflammatory cells and contribute to the disease process.[77] Lee et al[78] showed that SPINK1 overexpression was associated with liver cancer and portal vein invasion, and intestinal flora might intervene in NAFLD through SPINK1. Studies have shown that disorders of intestinal microbiota can easily lead to liver diseases. Intestinal metabolites can enter the blood and reach the liver through the portal vein through the enterohepatic circulation, resulting in inflammatory response and causing liver cell damage.[79] For example, studies by Yamada et al.[80] have found that palmitic acid, a long-chain fatty acid produced by intestinal flora, can promote the increase of CCL2 secretion, leading to a significant increase in the number of macrophages in the liver and aggravating high-fat diet-induced mouse steatosis. However, this change will be reversed after the removal of intestinal flora. These results suggest that some metabolites of intestinal flora can affect the process of NAFLD through macrophages. Elce et al.[81] showed that the metabolites of intestinal flora can activate the NF-κB signaling pathway and promote the expression of CCL2, IL-6, and IL-1B genes. Chaassaing et al.[82] found that intestinal flora disorders can cause the microflora infiltrating the inner layer of intestinal mucosa, contacting epithelial cells, promoting inflammatory response, and further leading to IR. After the combination of LPS and LPS-binding proteins in the outer layer of intestinal flora cell wall, nuclear transcription factors NF-κB and FOS were activated through a series of reactions, and the mRNA expression of TNF, IL6 and IL1B was increased, which further mediated the production of TNF-α, IL-1β, and other pro-inflammatory factors.[83–86] The inflammatory factors can further affect the phosphorylation of insulin receptor substrates, interfere with glucose transporter expression, and eventually develop IR.[87] Insulin resistance and IL-6 promote and cause each other, which is an important cause of NAFLD.[88,89] C5AR1,[68,69] PTGS2[73], and pro-inflammatory factors such as IL-1B and IL-6[90,91] are involved in the sensitivity of liver to insulin through inflammatory signaling pathways and induce the formation of insulin resistance. CCL2,[36,37] FOS,[50] SPINK1[53–55] and PTGS2[71,72] in adipose tissue may increase insulin resistance. Insulin resistance is closely related to the formation of NAFLD, and changes in intestinal flora composition can improve insulin resistance.[92] Therefore, we speculated that IL6, IL1B, CCL2, and FOS may be key genes for the development and progression of NAFLD, which may contribute to the relationship between NAFLD and intestinal flora disorder mainly through inflammation and IR. SPINK1, C5AR1, and PTGS2 may be key genes for intestinal flora to interfere with NAFLD, possibly through endotoxin, insulin resistance, and inflammation intervention.
TNF signaling pathway, AGE-RAGE signaling pathway in diabetic complications, and NF-kappa B signaling pathway have been shown several times with good enrichment in all KEGG enrichment results. We believe that these 3 pathways are the key pathways for intestinal flora to treat NAFLD. AGE/RAGE activation can increase oxidative stress and trigger a series of inflammatory reactions, angiogenesis, fibrosis, thrombosis, cell proliferation, and apoptosis.[93] Japanese researchers reported that in the case of diabetes, serum AGE levels are positively correlated with insulin resistance and negatively correlated with adiponectin levels, which is an important biomarker to distinguish NASH from SS.[94] Fehrenbach et al[95] found that AGE/RAGE activation could increase the synthesis of reactive oxygen species, activate NF-kappa B signaling pathway in HSCs, and make them differentiate into fibroblasts. Activation of the NF-kappa B signaling pathway is an important inflammatory response mechanism, such as inflammatory bowel disease and inflammation during liver injury,[96–98] and is also associated with fatty liver.[99] LPS binds to toll-like receptors on the macrophage surface and activates IKK, which phosphorylates or degrades I-κB. Free NF-κB enters the nucleus and promotes the transcription of iNOS and inflammatory cytokines such as TNF-α, IL-1β, and IL-6.[100,101] Literature studies have shown that NF-κB and TNF signaling pathways are important inflammatory pathways during cholestatic liver injury.[102] Alexander et al. found that the infiltration of neutrophils and inflammatory macrophages in Nlrp3 mutant mice depends on TNF signaling, which can improve LPS-driven liver injury, prevent the activation of hepatic stellate cells in Nlrp3 mutant mice, and trigger liver inflammation.[103] The pathogenesis of many chronic liver diseases, such as viral hepatitis, alcoholic liver disease, and explosive liver failure, is closely related to the dysfunction of the TNF signaling pathway.[104] Activation of the TNF signaling pathway may trigger NASH and liver fibrosis.[105] Therefore, we speculated that TNF signaling pathway, AGE-RAGE signaling pathway in diabetic complications, and NF-kappa B signaling pathway may play an important role in the intervention of intestinal flora in the occurrence and development of NAFLD.
In summary, we believe that the pathogenesis of NAFLD is mainly related to insulin resistance and inflammation. The main mechanism by which intestinal flora interferes with the pathogenesis of NAFLD is to inhibit the expression of inflammatory genes in the related pathways and reduce insulin resistance, thereby reducing NAFLD. Although our research discussed the mechanism of intestinal flora interferes with NAFLD, there are still some limitations. First, the research data comes from existing databases, so the authenticity and completeness of the results depend on the data. Second, the results do not reflect all the genuine cellular network characteristics in the organism, so further experiments will be needed to confirm the data in the future.
5. Conclusion
In this study, bioinformatics was used to explore the relationship between intestinal flora disorder and NAFLD, and the mechanism of intestinal flora interfering with NAFLD was discussed. Through Merge function intestinal flora disorders PPI network and NAFLD PPI network, we have acquired 20 potential targets for treating SS by intestinal flora, 7 for treating NASH, and 7 for treating NASH cirrhosis. The intestinal flora disorder PPI network was merged with the merge network of NAFLD progress through the Merge function of Cytoscape 3.7.2 software. We obtained 5 potential targets for intestinal flora to intervene in the progression of NAFLD from SS to NASH, 3 potential targets for intestinal flora to intervene in the progression of NAFLD from NASH to NASH cirrhosis, and 7 potential targets for intestinal flora to intervene in the progression of NAFLD from SS to NASH cirrhosis. Finally, the 7 targets (CCL2, IL6, IL1B, FOS, PTGS2, SPINK1, and C5AR1) were selected as the core potential targets for intestinal flora to intervene NAFLD. CCL2, IL6, IL1B, and FOS are mainly related to the occurrence and development mechanism of NAFLD, while PTGS2, SPINK1, and C5AR1 are mainly related to the intervention of intestinal flora in the occurrence and development of NAFLD. GO enrichment analysis showed that the gene functions of intestinal flora interfering with NAFLD are mainly reflected in basic biological processes, including regulating metabolic processes, epithelial development, and affecting immunity, such as positive regulation of reactive oxygen species metabolic process, acute inflammatory response, and external side of the plasma membrane. KEGG enrichment analysis showed that the pathway through which intestinal flora interfered with NAFLD was mainly closely related to signal transduction, immune regulation, and physiological metabolism. TNF signaling pathway, AGE-RAGE signaling pathway in diabetic complications, and NF-kappa B signaling pathway are the main pathways. Although our research discussed the mechanism, there are still some limitations. First, research data comes from existing databases, so the authenticity and completeness of the results depend on the data. Second, the results do not reflect all the genuine cellular network characteristics in the organism, so further experiments will be needed to confirm the data in the future.
In conclusion, we predicted that the intervention process of intestinal flora in NAFLD was mainly related to inflammatory response and AGE/RAEG signal transduction. Interfering with these mechanisms of intestinal flora may lead to the goal of curing NAFLD. However, as our study is based on data analysis, further experiments are needed to confirm this result. The preliminary results of this study confirmed the intervention role and related mechanisms of intestinal flora in the occurrence and development of NAFLD, laying a good foundation for further exploration of its mechanism of action.
Author contributions
Conceptualization: YL and JW; Data Curation and Software: XL; Methodology: WZ and JZ; Validation: SG and SJ; Investigation: HW and JL; Formal analysis: YT; Writing - Original Draft: YL; Supervision: JW. All authors read and approved the final version of the manuscript.
Supplementary Material
Abbreviations:
- AP-1 =
- transcription factor subunit
- C5AR1 =
- complement C5a receptor 1
- CCL2 =
- C-C motif chemokine ligand 2
- FOS =
- Fos proto-oncogene
- IL1B =
- interleukin 1 beta
- IL6 =
- interleukin 6
- NAFLD =
- nonalcoholic fatty liver disease
- NASH cirrhosis =
- Non-alcoholic steatohepatitis associated cirrhosis
- NASH =
- nonalcoholic steatohepatitis
- PTGS2 =
- prostaglandin-endoperoxide synthase 2
- SPINK1 =
- serine peptidase inhibitor Kazal type 1
- SS =
- simple steatosis
This work was supported by the Young Scientists Training Program of Beijing University of Chinese Medicine, State Key Laboratory of Generic Manufacture Technology of Chinese Traditional Medicine (Grant nos. 2010DQ740377), and the National Nature Science Foundation of China (Grant nos. 81673829).
The current analysis does not require ethical approval, because our analysis only collects uploaded data information from the public database search. The program does not process any patient’s personal data and will not cause any patient hurt.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors have no conflicts of interest to disclose.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. All data obtained or analyzed during this study are available from the published article and its supplementary information files. The datasets during the current study are available from the corresponding author upon reasonable request.
Supplemental Digital Content is available for this article.
How to cite this article: Liu Y, Liu X, Zhou W, Zhang J, Wu J, Guo S, Jia S, Wang H, Li J, Tan Y. Integrated bioinformatics analysis reveals potential mechanisms associated with intestinal flora intervention in nonalcoholic fatty liver disease. Medicine 2022;101:36(e30184).
Contributor Information
Yingying Liu, Email: 13226603346@163.com.
Xinkui Liu, Email: lxkchuige@163.com.
Wei Zhou, Email: weizhou19940530@163.com.
Jingyuan Zhang, Email: lindajyz@163.com.
Siyu Guo, Email: gsiyu1995@163.com.
Shanshan Jia, Email: cristielove@163.com.
Haojia Wang, Email: wanghaojia123@yeah.net.
Jialin Li, Email: superlin2019@163.com.
Yingying Tan, Email: 13226603346@163.com.
References
- [1].Younossi ZM, Marchesini G, Pinto-Cortez H, et al. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: implications for liver transplantation. Transplantation. 2019;103:22–7. [DOI] [PubMed] [Google Scholar]
- [2].Perumpail BJ, Khan MA, Yoo ER, et al. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World J Gastroenterol. 2017;23:8263–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Arab JP, Dirchwolf M, Álvares-da-Silva MR, et al. Latin American association for the study of the liver (ALEH) practice guidance for the diagnosis and treatment of non-alcoholic fatty liver disease. Ann Hepatol. 2020;19:674–90. [DOI] [PubMed] [Google Scholar]
- [4].Gadiparthi C, Spatz M, Greenberg S, et al. NAFLD Epidemiology, emerging pharmacotherapy, liver transplantation implications and the trends in the United States. J Clin Transl Hepatol. 2020;8:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- [6].Jumpertz R, L.e DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Neis EP, Dejong CH, Rensen SS. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sharon G, Garg N, Debelius J, et al. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014;20:719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marshall JC. The gut as a potential trigger of exercise-induced inflammatory responses. Can J Physiol Pharmacol. 1998;76:479–84. [DOI] [PubMed] [Google Scholar]
- [11].Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–9. [DOI] [PubMed] [Google Scholar]
- [12].Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pais R, Charlotte F, Fedchuk L, et al. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J Hepatol. 2013;59:550–6. [DOI] [PubMed] [Google Scholar]
- [14].Calzadilla Bertot L, Adams LA. The natural course of non-alcoholic fatty liver disease. Int J Mol Sci . 2016;17:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Arab JP, Karpen SJ, Dawson PA, et al. Bile acids and nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Hepatology. 2017;65:350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang H, Qin C, Li YH, et al. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim J, So S, Lee HJ, et al. DigSee: disease gene search engine with evidence sentences (version cancer). Nucleic Acids Res. 2013;41(Web Server issue):W510–W517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Piñero J, Bravo A, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Franz M, Lopes CT, Huck G, et al. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59:328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Poeta M, Pierri L, Vajro P. Gut-liver axis derangement in non-alcoholic fatty liver disease. Children (Basel). 2017;4:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72:558–77. [DOI] [PubMed] [Google Scholar]
- [28].Kwon HJ, Won YS, Park O, et al. Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty liver but worsens liver inflammation and fibrosis in mice. Hepatology. 2014;60:146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Caricilli AM, Saad MJ. The role of gut microbiota on insulin resistance. Nutrients. 2013;5:829–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fuchs C, Claudel T, Trauner M. Bile acid-mediated control of liver triglycerides. Semin Liver Dis. 2013;33:330–42. [DOI] [PubMed] [Google Scholar]
- [31].Blaut M. Gut microbiota and energy balance: role in obesity. Proc Nutr Soc. 2015;74:227–34. [DOI] [PubMed] [Google Scholar]
- [32].Camp JG, Jazwa AL, Trent CM, et al. Intronic cis-regulatory modules mediate tissue-specific and microbial control of Angptl4/Fiaf transcription. PLoS Genet. 2012;8:e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sierra-Filardi E, Nieto C, Domínguez-Soto A, et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. J Immunol. 2014;192:3858–67. [DOI] [PubMed] [Google Scholar]
- [34].Baeck C, Wei X, Bartneck M, et al. Pharmacological inhibition of the chemokine C-C motif chemokine ligand 2 (monocyte chemoattractant protein 1) accelerates liver fibrosis regression by suppressing Ly-6C(+) macrophage infiltration in mice. Hepatology. 2014;59:1060–72. [DOI] [PubMed] [Google Scholar]
- [35].Miura K, Yang L, van Rooijen N, et al. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Egan CE, Daugherity EK, Rogers AB, et al. CCR2 and CD44 promote inflammatory cell recruitment during fatty liver formation in a lithogenic diet fed mouse model. PLoS One. 2013;8:e65247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kamada Y, Kiso S, Yoshida Y, et al. Estrogen deficiency worsens steatohepatitis in mice fed high-fat and high-cholesterol diet. Am J Physiol Gastrointest Liver Physiol. 2011;301:G1031–43. [DOI] [PubMed] [Google Scholar]
- [38].Fenkci S, Rota S, Sabir N, et al. Relationship of serum interleukin-6 and tumor necrosis factor alpha levels with abdominal fat distribution evaluated by ultrasonography in overweight or obese postmenopausal women. J Investig Med. 2006;54:455–60. [DOI] [PubMed] [Google Scholar]
- [39].Anty R, Lemoine M. Liver fibrogenesis and metabolic factors. Clin Res Hepatol Gastroenterol. 2011;35(Suppl 1):S10–20. [DOI] [PubMed] [Google Scholar]
- [40].Kumar R, Prakash S, Chhabra S, et al. Association of pro-inflammatory cytokines, adipokines & oxidative stress with insulin resistance & non-alcoholic fatty liver disease. Indian J Med Res. 2012;136:229–36. [PMC free article] [PubMed] [Google Scholar]
- [41].Georgoulis M, Kontogianni MD, Tileli N, et al. The impact of cereal grain consumption on the development and severity of non-alcoholic fatty liver disease. Eur J Nutr. 2014;53:1727–35. [DOI] [PubMed] [Google Scholar]
- [42].Cengiz M, Yasar DG, Ergun MA, et al. The role of interleukin-6 and interleukin-8 gene polymorphisms in non-alcoholic steatohepatitis. Hepat Mon. 2014;14:e24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tilg H, Moschen AR. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol Metab. 2008;19:371–9. [DOI] [PubMed] [Google Scholar]
- [44].Dinarello CA, Donath MY, Mandrup-Poulsen T. Role of IL-1beta in type 2 diabetes. Curr Opin Endocrinol Diabetes Obes. 2010;17:314–21. [DOI] [PubMed] [Google Scholar]
- [45].Miura K, Kodama Y, Inokuchi S, et al. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology. 2010;139:323–34.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nozaki Y, Saibara T, Nemoto Y, et al. Polymorphisms of interleukin-1 beta and beta 3-adrenergic receptor in Japanese patients with nonalcoholic steatohepatitis. Alcohol Clin Exp Res. 2004;28:106S–10S. [DOI] [PubMed] [Google Scholar]
- [47].Woods A, Brull DJ, Humphries SE, et al. Genetics of inflammation and risk of coronary artery disease: the central role of interleukin-6. Eur Heart J. 2000;21:1574–83. [DOI] [PubMed] [Google Scholar]
- [48].Kanduri C, Raman R. Characterisation of developmentally regulated chromatin structure in the coding region of the proto-oncogene, c-fos, in the male laboratory mouse. Int J Dev Biol. 1999;43:279–82. [PubMed] [Google Scholar]
- [49].Dorn C, Engelmann JC, Saugspier M, et al. Increased expression of c-Jun in nonalcoholic fatty liver disease. Lab Invest. 2014;94:394–408. [DOI] [PubMed] [Google Scholar]
- [50].Jones MR, Chazenbalk G, Xu N, et al. Steroidogenic regulatory factor FOS is underexpressed in polycystic ovary syndrome (PCOS) adipose tissue and genetically associated with PCOS susceptibility. J Clin Endocrinol Metab. 2012;97:E1750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Videla LA, Tapia G, Rodrigo R, et al. Liver NF-kappaB and AP-1 DNA binding in obese patients. Obesity (Silver Spring). 2009;17:973–9. [DOI] [PubMed] [Google Scholar]
- [52].Hass HG, Jobst J, Vogel U, et al. Overexpression of tumor-associated trypsin inhibitor (SPINK1/TATI) in hepatitis C-associated hepatocellular carcinoma: potential implications for viral hepatocarcinogenesis. Oncol Res Treat. 2014;37:732–8. [DOI] [PubMed] [Google Scholar]
- [53].Itkonen O, Stenman UH. TATI as a biomarker. Clin Chim Acta. 2014;431:260–9. [DOI] [PubMed] [Google Scholar]
- [54].Averbukh LD, Mavilia MG. SPINK-1 Polymorphism as a pancreatitis risk factor. Cureus. 20192019;11:e3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wapenaar MC, Monsuur AJ, Poell J, et al. The SPINK gene family and celiac disease susceptibility. Immunogenetics. 2007;59:349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wiksten JP, Lundin J, Nordling S, et al. High tissue expression of tumour-associated trypsin inhibitor (TATI) associates with a more favourable prognosis in gastric cancer. Histopathology. 2005;46:380–8. [DOI] [PubMed] [Google Scholar]
- [57].Soreide K, Janssen EA, Körner H, et al. Trypsin in colorectal cancer: molecular biological mechanisms of proliferation, invasion, and metastasis. J Pathol. 2006;209:147–56. [DOI] [PubMed] [Google Scholar]
- [58].Higashiyama M, Monden T, Tomita N, et al. Expression of pancreatic secretory trypsin inhibitor (PSTI) in colorectal cancer. Br J Cancer. 1990;62:954–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ogawa M, Matsuura N, Higashiyama K, et al. Expression of pancreatic secretory trypsin inhibitor in various cancer cells. Res Commun Chem Pathol Pharmacol. 1987;55:137–40. [PubMed] [Google Scholar]
- [60].Ozaki N, Ohmuraya M, Hirota M, et al. Serine protease inhibitor Kazal type 1 promotes proliferation of pancreatic cancer cells through the epidermal growth factor receptor. Mol Cancer Res. 2009;7:1572–81. [DOI] [PubMed] [Google Scholar]
- [61].Oruc N, Ozutemiz O, Berdeli A, et al. Common SPINK-1 mutations do not predispose to the development of non-alcoholic fatty liver disease. Ann Hepatol. 2009;8:116–9. [PubMed] [Google Scholar]
- [62].Ohmachi Y, Murata A, Matsuura N, et al. Specific expression of the pancreatic-secretory-trypsin-inhibitor (PSTI) gene in hepatocellular carcinoma [published correction appears in Int J Cancer 1994 Apr 1;57(1):139]. Int J Cancer. 1993;55:728–34. [DOI] [PubMed] [Google Scholar]
- [63].Gerard NP, Gerard C. The chemotactic receptor for human C5a anaphylatoxin. Nature. 1991;349:614–7. [DOI] [PubMed] [Google Scholar]
- [64].Markiewski MM, DeAngelis RA, Benencia F, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9:1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Markiewski MM, Vadrevu SK, Sharma SK, et al. The ribosomal protein s19 suppresses antitumor immune responses via the complement C5a receptor 1. J Immunol. 2017;198:2989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Markiewski MM, Lambris JD. Unwelcome complement. Cancer Res. 2009;69:6367–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol. 2007;171:715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Orrem HL, Nilsson PH, Pischke SE, et al. IL-6 Receptor inhibition by tocilizumab attenuated expression of C5a receptor 1 and 2 in non-ST-elevation myocardial infarction. Front Immunol. 2018;9:2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schmitt J, Roderfeld M, Sabrane K, et al. Complement factor C5 deficiency significantly delays the progression of biliary fibrosis in bile duct-ligated mice. Biochem Biophys Res Commun. 2012;418:445–50. [DOI] [PubMed] [Google Scholar]
- [70].Sendler M, Beyer G, Mahajan UM, et al. Complement component 5 mediates development of fibrosis, via activation of stellate cells, in 2 mouse models of chronic pancreatitis. Gastroenterology. 2015;149:765–76.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dieter P, Scheibe R, Jakobsson PJ, et al. Functional coupling of cyclooxygenase 1 and 2 to discrete prostanoid synthases in liver macrophages. Biochem Biophys Res Commun. 2000;276:488–92. [DOI] [PubMed] [Google Scholar]
- [72].Enomoto N, Ikejima K, Yamashina S, et al. Kupffer cell-derived prostaglandin E(2) is involved in alcohol-induced fat accumulation in rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;279:G100–6. [DOI] [PubMed] [Google Scholar]
- [73].Vickers AE. Characterization of hepatic mitochondrial injury induced by fatty acid oxidation inhibitors. Toxicol Pathol. 2009;37:78–88. [DOI] [PubMed] [Google Scholar]
- [74].Dieter P, Scheibe R, Kamionka S, et al. LPS-induced synthesis and release of PGE2 in liver macrophages: regulation by CPLA2, COX-1, COX-2, and PGE2 synthase. Adv Exp Med Biol. 2002;507:457–62. [DOI] [PubMed] [Google Scholar]
- [75].Kosumi K, Hamada T, Zhang S, et al. Prognostic association of PTGS2 (COX-2) over-expression according to BRAF mutation status in colorectal cancer: Results from two prospective cohorts and CALGB 89803 (Alliance) trial. Eur J Cancer. 2019;111:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Loo TM, Kamachi F, Watanabe Y, et al. Gut microbiota promotes obesity-associated liver cancer through pge2-mediated suppression of antitumor immunity. Cancer Discov. 2017;7:522–38. [DOI] [PubMed] [Google Scholar]
- [77].Woodruff TM, Arumugam TV, Shiels IA, et al. A potent human C5a receptor antagonist protects against disease pathology in a rat model of inflammatory bowel disease. J Immunol. 2003;171:5514–20. [DOI] [PubMed] [Google Scholar]
- [78].Lee YC, Pan HW, Peng SY, et al. Overexpression of tumour-associated trypsin inhibitor (TATI) enhances tumour growth and is associated with portal vein invasion, early recurrence and a stage-independent prognostic factor of hepatocellular carcinoma. Eur J Cancer. 2007;43:736–44. [DOI] [PubMed] [Google Scholar]
- [79].Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72:558–77. [DOI] [PubMed] [Google Scholar]
- [80].Yamada S, Kamada N, Amiya T, et al. Gut microbiota-mediated generation of saturated fatty acids elicits inflammation in the liver in murine high-fat diet-induced steatohepatitis. BMC Gastroenterol. 2017;17:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Elce A, Amato F, Zarrilli F, et al. Butyrate modulating effects on pro-inflammatory pathways in human intestinal epithelial cells. Benef Microbes. 2017;8:841–7. [DOI] [PubMed] [Google Scholar]
- [82].Chassaing B, Raja SM, Lewis JD, et al. colonic microbiota encroachment correlates with dysglycemia in humans. Cell Mol Gastroenterol Hepatol. 2017;4:205–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15:1546–58. [DOI] [PubMed] [Google Scholar]
- [84].Gnauck A, Lentle RG, Kruger MC. The characteristics and function of bacterial lipopolysaccharides and their endotoxic potential in humans. Int Rev Immunol. 2016;35:189–218. [DOI] [PubMed] [Google Scholar]
- [85].Su GL, Klein RD, Aminlari A, et al. Kupffer cell activation by lipopolysaccharide in rats: role for lipopolysaccharide binding protein and toll-like receptor 4. Hepatology. 2000;31:932–6. [DOI] [PubMed] [Google Scholar]
- [86].von Montfort C, Beier JI, Guo L, et al. Contribution of the sympathetic hormone epinephrine to the sensitizing effect of ethanol on LPS-induced liver damage in mice. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wisniewski PJ, Dowden RA, Campbell SC. Role of dietary lipids in modulating inflammation through the gut microbiota. Nutrients. 2019;11:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bedi O, Aggarwal S, Trehanpati N, et al. Molecular and pathological events involved in the pathogenesis of diabetes-associated nonalcoholic fatty liver disease. J Clin Exp Hepatol. 2019;9:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Deng ZB, Poliakov A, Hardy RW, et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58:2498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Oliver E, McGillicuddy F, Phillips C, et al. The role of inflammation and macrophage accumulation in the development of obesity-induced type 2 diabetes mellitus and the possible therapeutic effects of long-chain n-3 PUFA. Proc Nutr Soc. 2010;69:232–43. [DOI] [PubMed] [Google Scholar]
- [91].Schulze F, Wehner J, Kratschmar DV, et al. Inhibition of IL-1beta improves glycaemia in a mouse model for gestational diabetes. Sci Rep. 2020;10:3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–81. [DOI] [PubMed] [Google Scholar]
- [93].Ott C, Jacobs K, Haucke E, et al. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014;2:411–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hyogo H, Yamagishi S, Iwamoto K, et al. Elevated levels of serum advanced glycation end products in patients with non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2007;22:1112–9. [DOI] [PubMed] [Google Scholar]
- [95].Fehrenbach H, Weiskirchen R, Kasper M, et al. Up-regulated expression of the receptor for advanced glycation end products in cultured rat hepatic stellate cells during transdifferentiation to myofibroblasts. Hepatology. 2001;34:943–52. [DOI] [PubMed] [Google Scholar]
- [96].Wang X, Sun Y, Zhao Y, et al. Oroxyloside prevents dextran sulfate sodium-induced experimental colitis in mice by inhibiting NF-κB pathway through PPARγ activation. Biochem Pharmacol. 2016;106:70–81. [DOI] [PubMed] [Google Scholar]
- [97].Ma P, Ding YS, Xuan LL, et al. Anti-inflammatory effect of a resveratrol derivative 3,4,5-trimethoxy-4’,5’-dihydroxy-trans-stilbene (WL-09-5) via ROS-mediated NF-κB pathway. J Asian Nat Prod Res. 2016;18:1004–13. [DOI] [PubMed] [Google Scholar]
- [98].Sun LD, Wang F, Dai F, et al. Development and mechanism investigation of a new piperlongumine derivative as a potent anti-inflammatory agent. Biochem Pharmacol. 2015;95:156–69. [DOI] [PubMed] [Google Scholar]
- [99].Zhang T, Hu J, Wang X, et al. MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway. J Hepatol. 2019;70:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang J, Zhu R, Sun D, et al. Intracellular uptake of curcumin-loaded solid lipid nanoparticles exhibit anti-inflammatory activities superior to those of curcumin through the NF-κB signaling pathway. J Biomed Nanotechnol. 2015;11:403–15. [DOI] [PubMed] [Google Scholar]
- [101].Kim KS, Lee DS, Bae GS, et al. The inhibition of JNK MAPK and NF-κB signaling by tenuifoliside A isolated from polygala tenuifolia in lipopolysaccharide-induced macrophages is associated with its anti-inflammatory effect. Eur J Pharmacol. 2013;721:267–76. [DOI] [PubMed] [Google Scholar]
- [102].Li Z, Chen D, Jia Y, et al. Methane-rich saline counteracts cholestasis-induced liver damage via regulating the TLR4/NF-κB/NLRP3 inflammasome pathway. Oxid Med Cell Longev. 2019;2019:6565283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wree A, McGeough MD, Inzaugarat ME, et al. NLRP3 inflammasome driven liver injury and fibrosis: roles of IL-17 and TNF in mice. Hepatology. 2018;67:736–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Schwabe RF, Brenner DA. Mechanisms of liver injury. I. TNF-alpha-induced liver injury: role of IKK, JNK, and ROS pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G583–9. [DOI] [PubMed] [Google Scholar]
- [105].Wullaert A, Heyninck K, Beyaert R. Mechanisms of crosstalk between TNF-induced NF-kappaB and JNK activation in hepatocytes. Biochem Pharmacol. 2006;72:1090–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.