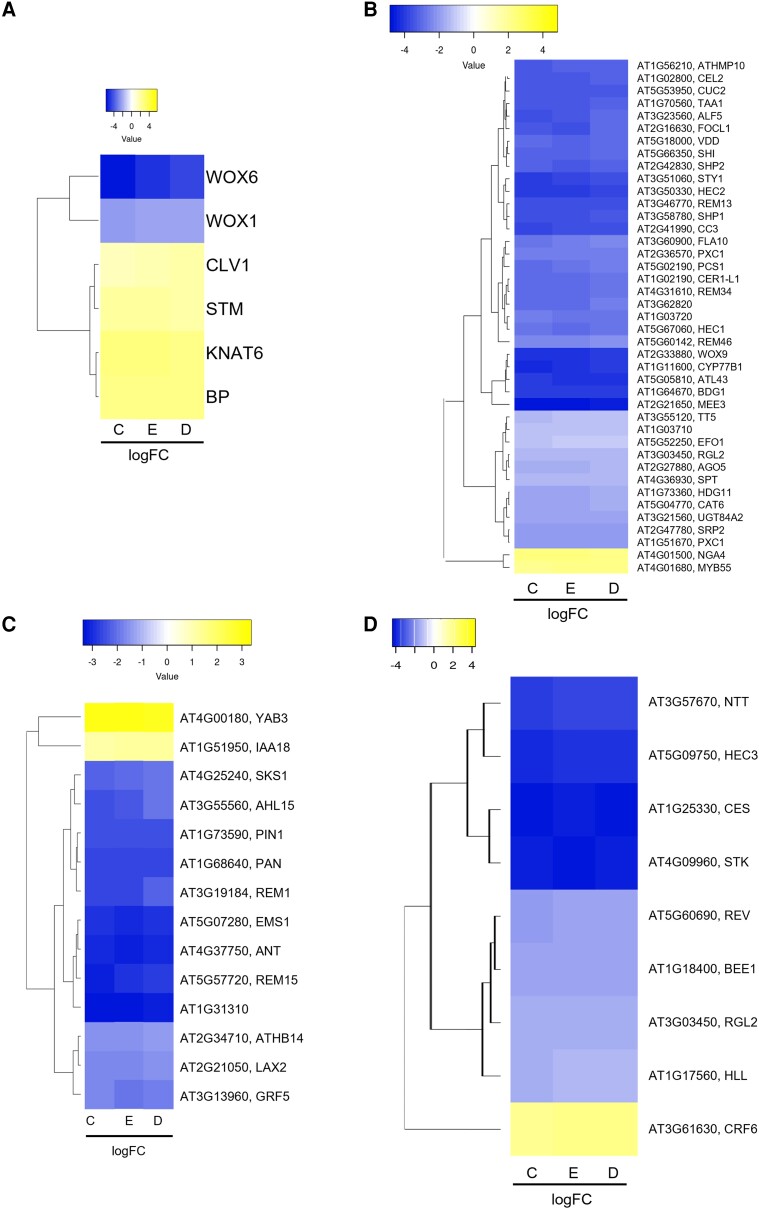
Figure 3.
CMM-specific genes are deregulated in the hda19-3 mutant. A) Meristem marker genes deregulated in the hda19-3 mutant. B) CMM-specific genes from stages 6 to 8 of pistil development according to Villarino et al. (2016) that are deregulated in the hda19-3 mutant. C) CMM-specific genes from stages 8 to 10 of pistil development according to Wynn et al. (2011) that are deregulated in hda19-3.D) Notable ovule number and TT regulators are deregulated in hda19-3. Genes plotted correspond to genes with an FDR < 0.001 and logFC > |1.2| in the RNA-Seq data according to Cufflinks (C), EdgeR (E), and DeSeq2 (D). Genes were hierarchically clustered by average clustering using Euclidean distances. Heatmaps were obtained using Heatmapper (http://heatmapper.ca/). The scales are relative to the maximum and minimum values of each table and represents the logFC (wild-type vs hda19-3) as detected by each program.