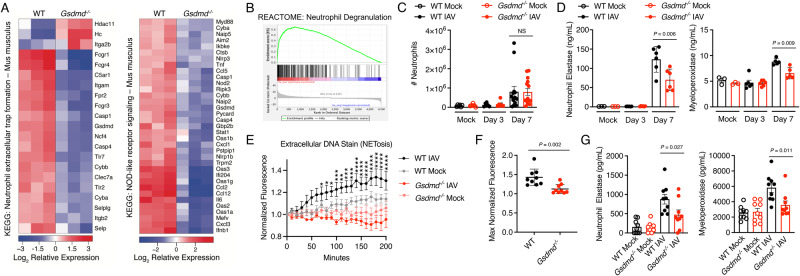
Fig. 5. IAV-induced neutrophil functions are decreased in the absence of GSDMD.
A RNAseq results as in Fig. 3 were analyzed to create heat maps for differentially regulated genes involved with neutrophil extracellular trap formation and NOD-like receptor signaling defined by KEGG pathway analysis. B REACTOME analysis of differentially expressed genes in WT versus Gsdmd−/− mice identified differences in genes associated with neutrophil degranulation. C Neutrophil infiltration into the lungs quantified by flow cytometry of mock-infected (from two independent experiments), day 3 post infection (from 1 experiment), or day 7 post infection (from 3 independent experiments) (each dot represents a single mouse, n = 6 mocks, n = 6 day 3, n = 14 day 7, error bars indicate SEM, NS, not significant by two-way ANOVA followed by Tukey’s multiple comparisons test). D ELISA quantification of neutrophil elastase and myeloperoxidase levels in lung homogenates of mock-infected (from 1 experiment), day 3 post infection (from 1 experiment), or day 7 post infection (from 2 independent experiments) (each dot represents a single mouse, n = 3 mocks, n = 6 day 3, n = 6 day 7, error bars indicate SEM, p values determined by two way ANOVA followed by Tukey’s multiple comparisons test). E Relative extracellular DNA from bone marrow neutrophils was quantified via fluorescence intensity readings following IAV infection (PR8, MOI = 10, each data point is an average of n = 9 with cells derived from 3 mice for each genotype assayed in triplicate, error bars indicate SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 determined by two-ANOVA followed by Bonferroni multiple comparisons test comparing WT IAV to Gsdmd−/− IAV, other comparisons not shown). F Maximum fluorescence intensity for each infected well as in E (p-value determined by two-tailed unpaired t-test). G Neutrophil products in supernatants from cells as in E were quantified via ELISA (n = 9 with cells derived from 3 mice assayed in triplicate, error bars indicate SEM, *p values determined by one-way ANOVA followed by Tukey’s multiple comparisons test). Source data are provided in the Source Data file.