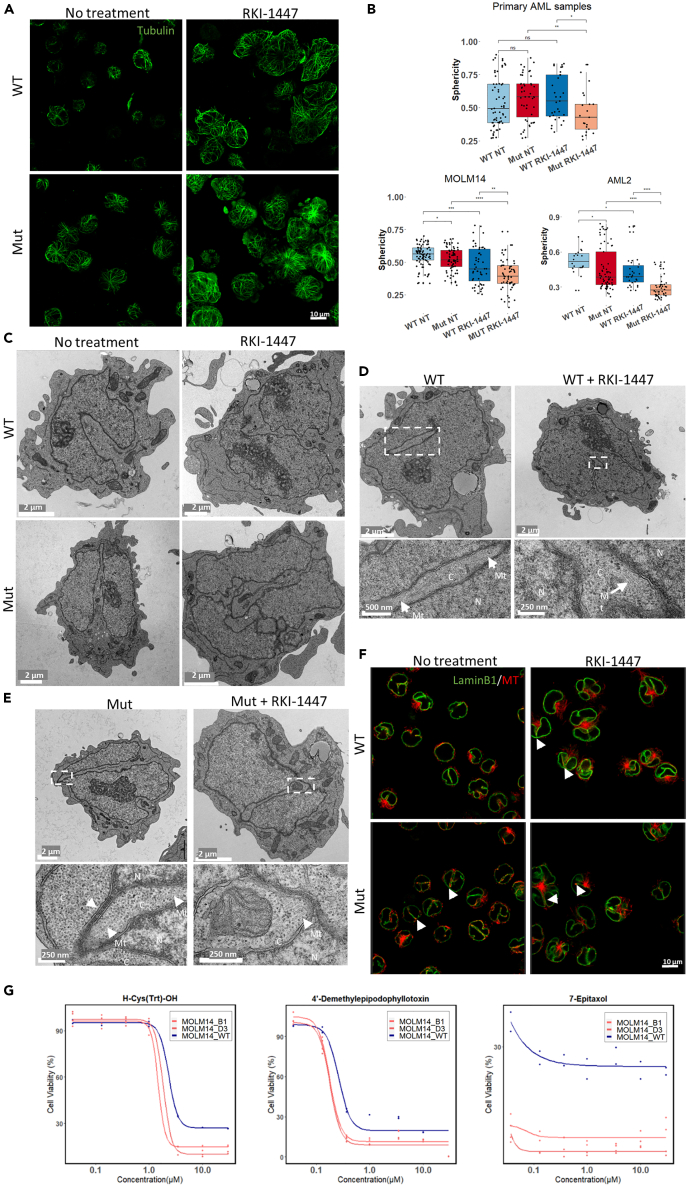
Figure 4.
Confocal and TEM images of SRSF2 WT/Mut cells before and after exposure to RKI-1447
SRSF2 WT/Mut MOLM14 and OCI-AML2 cells were either left untreated or treated with 0.5 μM RKI-1447 for 24 h. One SRSF2 Mut primary AML sample and one SRSF2 WT primary AML sample were either left untreated or treated with RKI-1447 (1 μM) for 48 h before fixation.
(A) Representative images of microtubules (shown in green), in SRSF2 WT/Mut MOLM14 cells, untreated or treated with RKI-1447 (0.5 μM), were acquired using the Leica SP8 scanning confocal microscope. Cells were labeled with anti-α-tubulin antibodies. A series of confocal slices depicting the overall morphology of the microtubular network in these cells is shown in Figure S8.
(B) 3D confocal images of DAPI-labeled nuclei were subjected to rendering and morphometric quantifications. Morphometric characteristics of rendered sphericity of primary AML samples (upper), MOLM14 cells, and AML2 cells (lower) are presented in box and whiskers plot format. Statistical analysis was performed using two-sided t tests; ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005.
(C) TEM showing a common presence of a deep (“half-way”) nuclear indentation (top-left image) in MOLM14 WT cells; In mutant MOLM14 cells, these nuclear indentations were considerably deeper, usually contacting the opposite side of the nucleus, thereby segmenting the nucleus (bottom-left image); treatment of WT cells with RKI-1447 induced conspicuous nuclear segmentation (top-right image). Note that segments are often connected to each other by extended sheets consisting of two nuclear membranes, inter-connected by the associated nuclear laminae; “hyper-segmentation” of the nuclei of the mutant cells (bottom-right). For higher magnification view, see also Figure S12.
(D and E) TEM images illustrate the overall morphology of SRSF2 WT (D) and mutant (E) MOLM14 cells, untreated or treated with RKI-1447 (0.5 μM) are shown in lower magnification (top panel) and higher magnification (bottom panel, corresponding to the white rectangles in the top panel). Nuclear regions and cytoplasmic regions are marked C and N, respectively. Arrows marked by “Mt” point to microtubules that are associated with the edges of the cytoplasmic insertions into the nucleus. The arrow in the “Mut” (bottom panel) points to the “interlobular sheet” consisting of the two nuclear membranes and the nuclear lamina running between them.
(F) Cells were labeled with anti-LaminB1 antibodies to outline nuclear membrane (shown in green) and anti-α-tubulin antibodies to label microtubules (shown in red). 3D volumes were acquired on the Olympus confocal microscope. Representative confocal slices corresponding to the middle plane of the cells are shown. Note deep narrow folds of nuclear membrane that are especially prominent in mutant cells and upon RKI-1447 treatment. Red signal detected inside such invaginations points to the presence of microtubules (arrows). See also Video S1 and Figure S14.
(G) Dose-response curves of three cytoskeleton modulators against SRSF2 WT and Mut MOLM14 (B1, D3) cell lines. Viability of SRSF2 WT and Mut MOLM14 cell lines was measured after 48 h exposure to three microtubule modifiers: H-Cys(Trt)-OH, 4′-Demethylepipodophyllotoxin and 7-Epitaxol at concentration of 0.037, 0.129, 0.369, 0.997, 3.49, 9.97, and 29.90 μM (see also Figure S17 for other MT modulators). The highest three concentrations were compared between WT and mutated clones by two-way ANOVA, followed by Dunnett’s post hoc test, see also Data S10.