Abstract
Fermentation is an effective means of enhancing the nutritional value of natural medicines, however, it is unclear how the metabolites changed during the fermentation of Paeonia lactiflora root (PLR). This study intends to elucidate how the active constituents and antioxidant activity of PLR change during fermentation. The study examined the levels of total glucosides of paeony (TGP), total flavonoids content (TFC), total phenols content (TPC), and antioxidant capability by high performance liquid chromatography (HPLC) and spectrophotometry. The chemical compositions before and after PLR fermentation were compared utilizing ultra-high performance liquid chromatography-mass spectrometry (UHPLC - MS). The findings from this study indicate that TGP, TFC and TPC peaked at Day 2 of fermentation, and the antioxidant capacity increased after fermentation. Of the 109 detected compounds, 18 were discrepant compounds. In summary, fermentation is an essential strategy for enhancing the functional activity of PLR. The current study could establish a scientific basis for future research on the fermentation of PLR, and provides new insights into the influence of fermentation on chemical composition as well as the antioxidant activity of drugs.
Keywords: Paeonia lactiflora root, Fermentation, UHPLC-ESI-Q-Orbitrap-MS, Differential metabolites analysis
1. Introduction
Paeonia lactiflora Root (PLR) has been prescribed in traditional Chinese medicine for medicinal prescriptions, and has excellent pharmacological effects and nutritional value [1]. The most important active substances in PLR are monoterpene glycosides, mainly including paeoniflorin, albiflorin, hydroxypaeoniflorin, and benzoylpaeoniflorin, which are generally referred to as total glucosides of paeony (TGP) [2], and are also rich in flavonoids, polyphenols and other active ingredients [3]. Therefore, PLR demonstrates exceptional antioxidant properties and is frequently utilized in treating conditions associated with oxidative stress, including arthritis, pneumonia, and fever [4,5]. However, the wild resources of Paeonia lactiflora have been seriously damaged, and the product have been cultivated for a long time. Thus, finding a method that can effectively enhance the content of the active components and the antioxidant capacity of PLR has become an important topic of the current research.
Microbial fermentation is one of the most commonly used methods for processing natural drugs. During the fermentation process, microorganisms can destroy the cell wall through enzymes such as cellulases and pectinases, which help to dissolve the active ingredients, and convert components including glycosides, phenolic acids, proteins into bioactive metabolites through glycosyl hydrolases, phenolic acid decarboxylases, and reductases, respectively, to enhance the pharmacological activity and nutritional value of the natural medicines [6,7]. In addition, during fermentation microorganisms produce enzymes with antioxidant activity such as superoxide dismutase and catalase, which can further enhance the antioxidant activity of natural medicines [8], making fermentation an ideal method for the treatment of natural medicines.
Fermentation is a complex process in which the chemical composition of the fermentation substrate changes considerably under the metabolic influence of the applied strain. Analyses of individual components often fail to fully interpret such changes [9]. Metabolomics offers a thorough examination of metabolites, especially of those that are undetectable using traditional techniques such as UV, as well as enable their identification and quantification [10]. Metabolomics technology has been used to examine the impact of fermentation strains, fermentation time, and raw materials on metabolites in fermented natural drugs and foods.
In this study, Saccharomycopsis fibuligera was selected for the fermentation of PLR. Metabolomics technology was applied to determine the compositional changes in small molecule compounds in PLR, identify the possible mechanism of action of microbial fermentation on the biotransformation of PLR, and explore the antioxidant effects of the fermented material. This research will provide actual data to support the development and application of PLR products derived from fermentation.
2. Materials and methods
2.1. Chemical reagents
The highly active and high cellulase-producing strain Saccharomycopsis fibuligera Y2 was obtained through the selection process from the original strain Saccharomycopsis fibuligera (CGMCC:2.5608) in our laboratory, Glucose (Tianjin Beifang Tianmedical Chemical Reagent Factory), Peptone (Beijing Aoboxing Biotechnology Limited Liability Company), Yeast Extract (OXOID, UK), anhydrous ethanol, potassium dihydrogen phosphate, acetic acid, isopropanol, 1,1-diphenyl-2- picrylhydrazyl (DPPH), sodium hydroxide, hydrochloric acid, salicylic acid, ferrous sulfate, sodium dihydrogen phosphate, hydrogen peroxide, trichloroacetic acid, pyrogallic gallic acid, and 2,2′-Azinobis-(3-ethylbenzthiazoline-6-sulphonate) (ABTS) were purchased from Tianjin Kaitong Chemical Reagent (Tianjing, China), Benzoic Acid, Quercetin, Gallic Acid were purchased from Chengdu Dexter Biotechnology (Chengdu, China).
2.2. Material preparation and fermentation
Yeast culture was performed using YPD medium consisting of 1% yeast extract, 2% peptone, and 2% glucose. Saccharomycopsis fibuligera was activated by incubating in YPD medium at 28 °C for 24h. PLR was purchased from Daxinganling, China. After drying at 50 °C, the samples were crushed, sieved, and subsequently mixed with distilled water (1:20, w/v). The mixture was then autoclaved at 121 °C for 20 min. Subsequently, 1% of Saccharomycopsis fibuligera was introduced into the mixture, which was then incubated at 28 °C for a duration of 5 days. PLR from day 0 was taken as Paeonia lactiflora root unfermented liquid (PLR-U) for comparison. After PLR-U and Paeonia lactiflora root fermented liquid (PLR-F) reached the incubation time, they were centrifuged (5030×g) for 10 min, filtered, and frozen at −18 °C for assay.
2.3. Determination of active ingredients
2.3.1. Determine total glucosides of paeony
Determination of TGP content in PLR-U and PLR-F was performed by alkaline hydrolysis - HPLC [11]. HPLC analysis was conducted using a HPLC 1260 Infinity II (Agilent Technologies, USA), and the C18 chromatographic column (250 mm × 4.6 mm, 5 μm, Elite Analytical Instruments Co., Dalian, China) was maintained at 35 °C, with 0.05 mol L−1 potassium dihydrogen phosphate-acetic acid (350:8, mobile phase A) and methanol-isopropanol (100:6, mobile phase B) as the mobile phase with a flow rate of 0.8 mL min−1, A:B = 70:30, and the detection wavelength utilized was set at 230 nm. Appropriate amount of fermentation broth was evaporated to dryness, and hydrolyzed by adding 10 mL 1% NaOH solution. 0.5 mL of sample solution was added into the mobile phase to be adjusted to 10 mL, and the sample was analyzed after passing through 0.22 μm microporous filter membrane. Peak area was quantified by external standard method, with benzoic acid serving as the standard (Fig. S2a), and the TGP content was evaluated using the following formula, where 480.27 is the molecular weight of paeoniflorin and 122.12 is the molecular weight of benzoic acid.
2.3.2. Determine total flavonoid content
TFC was measured by Chen et al. method with modifications [12]. The procedure was as follows: 0.3 mL of 5% NaNO2 (w/v) was mixed with 0.5 mL of an appropriately thinned sample. After 6 min, 0.3 mL 10% Al(NO3)3 (w/v) was dispensed. The sample was reacted for an additional 6 min, and 4.0 mL of 4% NaOH (w/v) was dispensed and mixed well. The volume was fixed to the scale, and incubated for 15 min. The wavelength for sample detection was set to 510 nm. TFC was determined using the external standard method and by establishing standard curves (Fig. S2b), and expressed as mg·g−1 of quercetin.
2.3.3. Determine total phenolic content
The Folin-Ciocalteu colorimetric assay was performed to quantify TPC [13]. Briefly, 50 μL of the correspondingly thinned samples were mixed with 0.5 mL of forint-phenol solution (0.5 M) and incubated for 3 min. Subsequently, 2 mL of Na2CO3 (10%, w/v) was introduced into the mixture. Following calibration based on the scale, the reaction was shielded from light for a duration of 30 min. The wavelength for sample detection was set to 765 nm. TPC was determined using the external standard method and by establishing standard curves (Fig. S2c), and expressed as mg·g−1 of gallic acid equivalent (GAE).
2.4. Measurement of antioxidant capacity
In this study, DPPH, ABTS, Hydroxy (OH) radical scavenging capacity, and total reducing power were used to compare the antioxidant activity of PLR-U and PLR-F. DPPH and ABTS+ radical scavenging capacities were determined by the method of Cirlini et al. [14]. 2 mL of the sample was mixed with an equal amount of DPPH (0.15 mM), and the absorbance at 517 nm was measured after 0.5 h of reaction in a light-free environment. The mathematical formula applied was as follows: Scavenging rate% = [1 - (Abssample − Abscontrol)/Absblank]*100. For the ABTS+ assay, 1 mL of the sample is mixed with 4 mL of the ABTS+ working solution and allowed to react for 6 min in a light-free environment. The absorbance at 734 nm was measured. The mathematical formula used was as follows: Scavenging rate% = [(Absblank - Abssample)/Absblank]*100. A control was established using sodium phosphate buffer in place of the DPPH or ABTS+ solution, while the sample substituted by sodium phosphate buffer as a blank. For the OH− scavenging capacity, 2 mL of FeSO4 (6 mM), salicylic acid (70% ethanol solution), fermentation broth, and H2O2 (6 mM) were added sequentially, mixed thoroughly and reacted at 37 °C for 30 min. Distilled water served as the control instead of salicylic acid, while it functioned as the blank instead of the sample [15]. The absorbance was recorded at 510 nm. The mathematical formula used was as follows: Scavenging rate% = [(Absblank - Abssample)/Absblank]*100. For the total antioxidant reduction capacity, 2 mL of fermentation broth, sodium phosphate buffer (pH 7.2), and 10% C6FeK3N6 solution were added, reacted at 50 °C for 20 min, and 10% C2HCl3O2 solution was added, then left for 10 min [16]. Centrifuge at 3000×g for 10 min, collect the supernatant, combine it with 0.4 mL of 0.1% FeCl3, incubate for 10 min, and measure the absorbance at 700 nm. The value of absorbance reflects the strength of the total antioxidant reduction capacity. Moreover, antioxidant potentials were expressed with the half-maximal effect concentration (EC50) and the concentration of the sample when the absorbance reaches 0.5 in the total reducing power assay (RP0.5AU) values. GraphPad Prism 8.0.2 was used to calculate EC50 and RP0.5AU values.
2.5. Metabolomics analysis
The fermentation broth was added to the pre-cooled acetonitrile/water solution (1:1, V/V) and vortexed. After sonication at 4 °C for 30 min, the mixed samples were centrifuged (12075×g, 4 °C) for 20 min. The liquid portion was used for metabolomics analysis. The PLR-U and PLR-F extracts (5 μL each) were injected into a UHPLC-ESI-Q-Orbitrap-MS (UPLC, Thermo Fisher, USA) equipped with a Thermo Fisher Science Hypersil Gold AQ C18 column (100 × 2.1 mm, 1.9 μm). The mobile phase was consisted of solvent A (water, 0.1% formic acid) and solvent B (acetonitrile, 0.1% formic acid). The mobile phase flow rate was 0.3 mL min−1. The gradient program was as follows: 0–1 min, 2% B, 1–5 min, 2%–5% B, 5–10 min, 12%–20% B, 10–12 min, 20%–30% B, 12–13 min, 30%–50% B, 13–15 min, 50%–100% B, 15–16 min, 100% B, 16–19 min, 5% B, 19–20 min, 2% B. The parameters of MS were set as follows: Sheath gas flow rate, 30 psi, Auxiliary gas flow rate, 10 psi, Sweep gas flow rate, 0 psi, Spray voltage, 3.5 kV. Spray voltage, 3.5 kV, Capilary temp, 320 °C, Aux gas heater temp, 350 °C, Collision gas: Nitrogen, Normalized collision energies, 20, 40, 60 eV, s-lens: 60.0. In conjunction with selected a primary mass spectrometry full scan, the secondary mass spectrometry scan mode is automatically triggered. The primary resolution was set to 70,000 full width at half maximum (FWHM), while the secondary resolution was 17,500 FWHM. The ion scanning range covered m/z 50–1500 with a cycle counting of 3 times. A four-stage isolation window of 1.5 m/z was utilized, and a dynamic exclusion time of 5 s was applied. Simultaneously, quality control (QC) samples were collected after every three samples to assess the stability of the instrument used for sample collection.
2.6. MS data analysis and statistical analysis
MS raw data were treated by Compound Discoverer 3.3 (Thermo Fisher Scientific, USA), which focuses on peaks alignment, peaks detection, features filtering and metabolites annotation. The retention time was configured at 0.2 min, and the mass tolerance was established at 5 ppm. A signalto-noise of 1.5 and a peak intensity threshold of 10,000 were selected for peak detection. Peaks in the sample with more than 80% missing values were removed during data filtering. Total ion current (TIC) normalizatio was conducted to reduce instrumental variation before statistical analysis. Metabolites were tentatively identified by matching their MS/MS data with entries in ChemSpider, mzCloud, and internal databases. The obtained quantitative data were centralized and autoscaled using MetaboAnalyst (https://www.metaboanalyst.ca/MetaboAnalyst), resulting in the reduction of discrimination between high-abundance and low-abundance chromatographic peaks. Principal Component Analysis (PCA) and heatmaps were created by utilizing MetaboAnalyst and R language (Ver. 4.2.2). Differential metabolites were determined by Log2FC ≥ 1, p < 0.05, and P-value was adjusted for false discovery rate. The pathway information for differential metabolites was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Results of the active component content and antioxidant capacity measurements are presented as mean ± standard deviation. Statistical Product and Service Solutions (SPSS) Ver. 16.0 was used for data analysis. An ANOVA was used to determine the statistical significance of the differences, followed by a post hoc Tukey test. Statistically significant differences were identified for p-values less than 0.05.
3. Results
3.1. Active ingredient content
Trends of TGP, TFC, and TPC during fermentation process were shown in Fig. 1a, and the TGP chromatogram is shown in Fig. 1b. The TGP and TFC of PLR-F decreased on day 1, increased on day 2 and then decreased; while TPC of PLR-F increased before day 2 and then decreased. At the common turning points on day 2, the TGP, TFC, and TPC elevated by 43,86%, 26.38%, and 57.91%, respectively (Fig. 1a). TPC was 28.88% higher than PLR-U at day 1, probably due to the better water solubility of polyphenols, which the accumulating metabolite could be dissolved and detected in the liquid culture. All three active components increased significantly on Day 2, which may due to the fact that with the vigorous growth, the microorganisms, exhibit high metabolic activity, and various enzymes (e.g., pectinase, cellulase, etc.) could be secreted and consumed components in the PLR, (e.g., starch, protein, pectin, etc.). This effectively destroyed the cell wall and led to the release of the active components in PLR. As the fermentation process continued, microbes used, destroyed, and altered the active chemicals in PLR, resulting in a decrease in their content. This phenomenon indicates that short-term fermentation can effectively enhance the content of various active ingredients within the PLR, and reached a peak at 2 d, so day 2 samples were chosen for the subsequent antioxidant capacity and metabolomics analysis.
Fig. 1.
Changes in active ingredient content of PLR during the fermentation process. (a) Changes in PLR active ingredient content during fermentation process (n = 3). Bars with distinct letters represent a statistically significant difference (p < 0.05). (b) The chromatogram of PLR during different fermentation periods.
3.2. Antioxidant capacity
The antioxidant capacity of natural medicine relies on the presence of distinct chemicals with varying modes of action [17,18]. Therefore, we evaluated the antioxidant effects of PLR-F using four methods: DPPH, ABTS+ and OH− scavenging capacity, as well as the total reducing power, EC50 and RP0.5AU values before and after PLR fermentation were shown in Table 1 and Fig. S3. Compared to PLR-U, PLR-F has significant lower EC50 values of DPPH, ABTS+, OH−, and the RP0.5AU values of total reducing power (p < 0.05). The findings suggest that PLR-F can achieve the same antioxidant capacity at a lower concentration than PLR-U. The study found that PLR-F has a stronger antioxidant potential than PLR-U.
Table 1.
EC50 values and RP0.5AU values of PLR antioxidant capacity before and after fermentation.
| Index | Antioxidant activity | PLR-U(μg/mL) | PLR-F(μg/mL) |
|---|---|---|---|
| EC50 | DPPH clearance rate | 25.47 ± 0.38 | 22.22 ± 0.19* |
| ABTS+ clearance rate | 204.43 ± 0.23 | 60.17 ± 0.11* | |
| OH− clearance rate | 151.57 ± 7.96 | 74.35 ± 1.59* | |
| RP0.5AU | total restoring power | 66.01 ± 0.87 | 50.56 ± 0.35* |
Data were present as the mean ± SD (n = 3). Asterisks (*) represent the significant difference when comparing the PLR-U (p < 0.05).
3.3. Metabolomics analysis
To explore fermentation changes, we applied non-targeted metabolomics on PLR-U and PLR-F (fermented for 2 days). Total ion chromatography (TIC) of PLR-U and PLR-F were shown in Supplementary Fig. 1. The TIC plot indicated notable variations in the metabolites of samples pre- and post-fermentation.
A total of 109 metabolites were discovered in the investigation, with 62 found in positive mode and 47 in negative mode (Supplementary Table 1). Fig. 2 shows heatmaps and related pie charts for these metabolites to help visualize their content and categorization. It includes 16 terpenoids, 13 amino acids and their derivatives, 14 phenolic, 24 flavonoids, 10 sugars, 15 organic acids, 9 fatty acids, 6 coumarins, and 2 others. Terpenoids, flavonoids, phenolic, organic acids, amino acids, and derivatives dominate PLR metabolites by quantity.
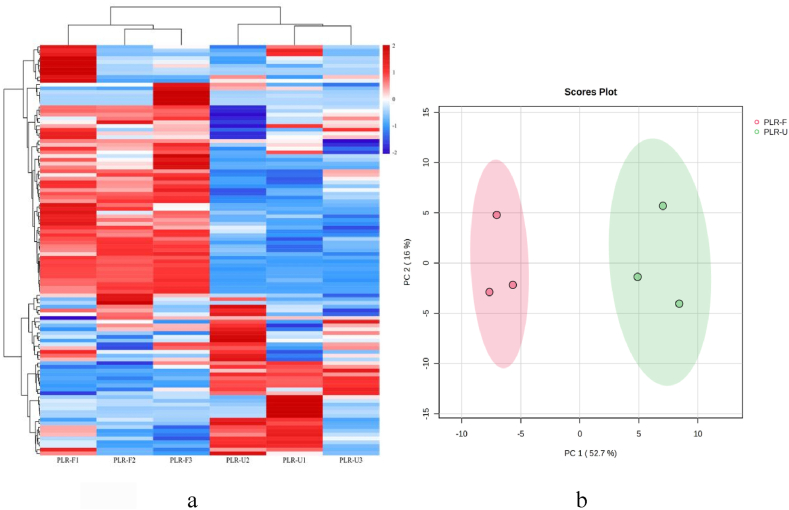
Fig. 2.
Heatmap visualization and proportions of the metabolites identified from PLR-U and PLR-F and multivariate analysis of identified metabolites. (a) Heatmap visualization. Metabolites with low relative abundance are represented in blue, whereas red denotes high abundance. (b) PCA analysis of metabolites identified from PLR-U and PLR-F. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Next, the differences in metabolite composition before and after fermentation were quantified by PCA. As seen in Fig. 2c, highly significant differentiation were shown between the two groups, indicating a high degree of cohesion and good segregation within the pre- and post-fermentation populations. This suggesting that fermentation can dramatically change the metabolites of PLR.
Volcano plots were created based on p < 0.05 and Log2FC ≥ 1 to illustrate differential metabolites (Fig. 3a). The findings demonstrated that in PLR-F, 5 compounds were considerably decreased and 13 compounds were significantly increased (Fig. 3b and c). Specific changes are shown in Table 2. In short, more metabolites were up-regulated after fermentation, suggesting that fermentation can effectively up-regulate various active components in PLR.
Fig. 3.
Differential metabolites between PLR-U and PLR-F. (a) A volcano graphic displaying the 109 differential metabolites that were found. (b) Pie chart showing metabolic divisions of PLR-U and PLR-F differential metabolites. (c) Differences in the alterations in various metabolites between PLR-U and PLR-F. (d) Differential metabolite KEGG pathway analysis. (e) Heatmap displaying the differential metabolites between PLR-F and PLR-U.
Table 2.
Differential metabolites of PLR-U and PLR-F were determined by UPLC-MS analysis.
| Classification | Compounds | Type | FC | Classification | Compounds | Type | FC |
|---|---|---|---|---|---|---|---|
| amino acids and derivatives | L-Serine | Up | 19.815 | flavone | 3,4,2′,3′,4′,6′,alpha-Heptahydroxychalcone 2′-glucoside | Down | 0.188 |
| L-Tryptophan | Up | 5.974 | (2S)-Naringenin | Up | 5.134 | ||
| L-Threonine | Up | 4.126 | Naringeninchalcone | Up | 3.392 | ||
| L-(+)-Aspartic acid | Down | 0.358 | 5,3′,4′,5′-Tetrahydroxy-6,7-dimethoxyflavone | Up | 3.296 | ||
| terpenoids | Mudanpioside J | Down | 0.179 | Catechin | Up | 2.801 | |
| plant polyphenol | Protocatechuic acid | Down | 0.209 | Quercetin | Up | 2.437 | |
| Protocatechuic aldehyde | Up | 2.176 | Naringenin trimethyl ether | Up | 2.429 | ||
| sugars | 1,3,6-Trigalloylglucose | Up | 2.985 | organic acids | Syringic acid | Up | 5.994 |
| lipids | (12Z)-9,10,11-trihydroxyoctadec-12-enoic acid | Up | 3.240 | Citric acid | Down | 0.385 |
*, metabolites detected in PLR-F but not in PLR-U. #, metabolites detected in PLR-U but not in PLR-F.
3.4. KEGG analysis
We conducted pathway enrichment analysis of the differential metabolites utilizing the KEGG database, and the outcomes are displayed in Fig. 3d. A total of 24 metabolic pathways were identified. Among the most important metabolic were glycine, serine and threonine metabolism, flavonoid biosynthesis, flavonoid and flavonol biosynthesis, cyanoamino acid metabolism, and glyoxylate and dicarboxylate metabolism.
3.5. Differences in chemical composition between PLR-U and PLR-F
In this experiment, the changes in chemical composition between PLR-U and PLR-F were analyzed, which laid the foundation for further study of PLR. Fig. 3e showed the heat map in the changes of different metabolites of PLR-U and PLR-F. Table 2 showed the specific information of 18 different compounds before and after the fermentation of PLR.
Table 2 and Fig. 3e show that differential metabolites included flavonoids, amino acids, polyphenols, terpenoids, organic acids, and fatty acids, suggesting that fermentation-induced changes in metabolites include a wide range of compound classes within the PLR. Flavonoids accounted for 38.9% and amino acids accounted for 22.2%, making them the main differential metabolites between PLR-F and PLR-U (Fig. 3b).
As can be seen from the figure, there was an increase in all flavonoids except 2-Hydroxyformononetin and 3,4,2′,3′,4′,6′, Alpha-Heptahydroxychalcone 2′-Glucoside. The KEGG study of differential metabolites indicated their primary involvement in flavonoid biosynthesis, as well as flavonoid and flavonol biosynthetic pathways. Catechin, Kaempferol, Quercetin, and (2S)-Naringenin are the important constituents of flavonoids in PLR, as can be seen from Table 2 and Table S2, the contents of Catechin, Kaempferol, Quercetin, (+)-Taxifolin and (2S)-Naringenin in PLR-F were increased by 2.80, 2.09, 2.44, and 5.13 folds compared to PLR-U, whereas the contents of the glycosidic forms such as 3,4,2′,3′,4′, 6′,Alpha-Heptahydroxychalcone 2′-Glucoside were decreased to some extent, indicating that the fermentation may have achieved a glycosidic effect. In addition, as can be seen from the table, except for L-(+)-Aspartic acid and L-Tyrosine, the levels of each other amino acids has increased. This increase may be attributed to microbial fermentation breaking down protein macromolecules in the PLR into smaller molecules like peptides and free amino acids. Furthermore, during fermentation, organic acids, and polyphenols accounted for 11.1% of the differential compounds and were the major compound species affected by fermentation.Among them, geranylacetone, syringic acid, and protocatechuic aldehyde are among the compounds up-regulated by fermentation and have been reported to exhibit good antioxidant activity. In summary, fermentation up-regulated the majority (72.2%) of differential metabolites, many of which exhibited strong antioxidant capacity. This accounts for the observed increase in PLR antioxidant activity following fermentation.
4. Discussion
PLR is a natural medicinal that is commonly utilized in compound formulas and medicinal diets for treating diseases and skin eruptions caused by heat substances invading the blood, menstrual closure and dysmenorrhea, and bruising and fluttering injuries [19]. Modern pharmacological studies have found that PLR is rich in monoterpene glycosides, flavonoids, polyphenols and amino acid compounds, which can exert antioxidant, anti-inflammatory and hepatoprotective effects with good application and nutritional value. However, at present, wild PLR resources have been seriously destructed in China. Because of special characteristics of its medicinal site (root), which often requires a growth period of more than three years and is a nonrenewable resource [20], a certain degree of scarcity exists in the market for PLR. Therefore, how to improveing the utilization rate of PLR resources is highly important for PLR development. Microbial fermentation improves plant nutrition, function, and pharmacological activity safely and effectively. Microbial fermentation produces a variety of hydrolases like exoglucanase, amylase, and hemicellulase, and simultaneously produces several secondary metabolites [21]. The treatment of PLR with microbial fermentation can effectively enhance the concentration of active compounds and contribute greatly to its pharmacological activity and nutritional value. Therefore, it is important to utilize appropriate bacterial strains for fermentation to broaden the value of PLR. The Mevalonic acid (MVA) pathway in the cytoplasm of plant cells has been reported to provide precursors for terpenoids, and yeasts have a highly similar MVA pathway. Compared to Escherichia coli, algae, and molds, yeast is considered more suitable for use as a chassis cell for terpenoid product synthesis [22,23]. Thus, Saccharomycopsis fibuligera was selected for the fermentation of PLR, and the results indicated enhanced terpenoid content in PLR. In addition, in our previous work, we examined the fermentation effects of various strains of Saccharomyces cerevisiae, Streptococcus thermophilus, and Aspergillus niger on PLR, and the results showed that Saccharomyces fibuligera had the most pronounced enhancement of various active ingredients in PLR.
The health benefits and therapeutic effects of fermented products in humans can be linked to changes in the active component makeup. Therefore, the integral characterization and description of small molecules in fermentation products are important to assess their active roles [24]. Most of the studies on PLR have concentrated on analyzing a particular active component, which typically does not offer a comprehensive perspective of its overall effectiveness. Metabolomics was used in this study to examine alterations in the metabolic profiles of PLR before and after fermentation for the first time. We observed notable variations between PLR-U and PLR-F based on metabolomics data and examined various main and secondary metabolites. Through differential metabolite analysis, it was discovered that fermentation primarily up-regulated flavonoids, including Catechin, Quercetin, and (2S)-Naringenin. These flavonoids have been reported to possess strong antioxidant activity, which explains the observed increase in overall antioxidant activity resulting from fermentation [[25], [26], [27], [28]]. We found that most flavonoids present in the form of glycosides were downregulated, suggesting that the glycosides were converted to more active glycosidic forms through the action of enzymes [29]. Furthermore, the levels of L-Serine, L-Tryptophan, and L-Threonine in PLR-F were found to be significantly increased. This could be attributed to the metabolic activity of the strains, leading to the breakdown of large proteins into smaller amino acids [30], and these findings are in line with the results obtained from the KEGG analysis. These additional amino acids not only possess excellent antioxidant capacity but also effectively enhance the nutritional value of PLR [[31], [32], [33]]. Tyrosine is essential for coumarins and flavonoids [34]. The decrease in tyrosine levels observed in PLR-F may be attributed to its conversion into flavonoids and coumarins during fermentation. After comparing these results with those of previous studies, we found that most of the active ingredients exhibited good activity. This may be the primary reason for the strong activity of PLR-F. This study demonstrates the innovative use of fermentation to significantly boost the active ingredient content and antioxidant capability of PLR. The findings suggest that PLR-F holds tremendous promise for the development of novel functional products, offering enhanced health benefits to consumers.
5. Conclusion
In summary, we used Saccharomycosis fibuligera Y2 for PLR fermentation, and dynamically measured the levels of TGP, TFC, and TPC and tested their antioxidant capacity during the fermentation process. The improvement of antioxidant capacity upon fermentation could be supported by altered metabolites composition between PLR-U and PLR-F from metabolomics analysis. Specifically, the study found that the TGP, TFC, and TPC contents within PLR were most significantly elevated at Day 2 of fermentation, therefore, PLR-F fermented for 2 d was selected for subsequent experiments. Compared to PLR-U, PLR-F showed superior antioxidant capacity. In this work, the chemical makeup of PLR both before and after fermentation was examined using metabolomics for the first time. The results demonstrated that fermentation primarily increased the flavonoid content. Additionally, there was also a noticeable increase in the levels of amino acids, polyphenols, and terpenes. KEGG analysis showed that the biosynthesis of flavonoids and flavonols, aminoacyl-tRNA, and metabolism of glycine, serine, and threonine were highly enriched metabolic pathways. In summary, this study explains the differences in the metabolite content and antioxidant activity between PLR-U and PLR-F. These results indicated that fermentation is an effective strategy for improving PLR activity and enhancing its utilization. Moreover, the remarkable antioxidant capacity exhibited by the fermentation products of PLR highlights their potential for application in treating diseases induced by oxidative stress. The study presents an innovative method for the biotransformation of natural compounds and establishes a conceptual framework for the future exploitation of PLR. The next work requires further research, such as purification of the fermentation products and further bioactivity verification.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Tianyu Wang: Writing – original draft, Data curation, Conceptualization. Kairui Sheng: Software, Investigation. Yifan Zhang: Visualization, Investigation. Songlin Jin: Validation. Linlin Feng: Methodology, Investigation, Conceptualization. Lihong Wang: Writing – review & editing, Resources, Project administration, Funding acquisition, Formal analysis.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Wang Lihong reports financial support was provided by Department of Education of Heilongjiang Province, China. The other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the “Jiamusi University Science and Technology Innovation Team School-level Innovation Teams” (Grant number: cxtd202103) and “North Medicine and Functional Food Characteristic Subject Project in Heilongjiang Province” (Grant number: HLJTSXK-2022-03).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28450.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ye X.W., Xia L.T., Ren H.M., et al. Progress in the historical development of white peony concoction and research on chemical composition and pharmacological effects. Chinese Herbal Medicine. 2020;51(7):1951–1969. [Google Scholar]
- 2.Zhang L., Wei W. Anti-inflammatory and immunoregulatory effects of paeoniflorin and total glucosides of paeony. Pharmacol. Therapeut. 2020;207 doi: 10.1016/j.pharmthera.2019.107452. [DOI] [PubMed] [Google Scholar]
- 3.Zhao D., Tao J., Han C., et al. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.) Mol. Biol. Rep. 2012;39(12):11263–11275. doi: 10.1007/s11033-012-2036-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang J.-M., Sun Y., Wang M., et al. Regulatory effect of a Chinese herbal medicine formula on non-alcoholic fatty liver disease. World J. Gastroenterol. 2019;25(34):5105–5119. doi: 10.3748/wjg.v25.i34.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad M., Malik K., Tariq A., et al. Botany, ethnomedicines, phytochemistry and pharmacology of Himalayan paeony (Paeonia emodi Royle) J. Ethnopharmacol. 2018;220:197–219. doi: 10.1016/j.jep.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Shakya S., Danshiitsoodol N., Sugimoto S., et al. Anti-oxidant and anti-inflammatory substance generated newly in Paeoniae Radix Alba extract fermented with plant-derived Lactobacillus brevis 174A. Antioxidants. 2021;10(7):1071. doi: 10.3390/antiox10071071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balcázar-Zumaeta C.R., Castro-Alayo E.M., Cayo-Colca I.S., et al. Metabolomics during the spontaneous fermentation in cocoa (Theobroma cacao L.): an exploraty review. Food Res. Int. 2023;163 doi: 10.1016/j.foodres.2022.112190. [DOI] [PubMed] [Google Scholar]
- 8.Yang J.Y., Kim G.R., Chae J.S., et al. Antioxidant and anti-inflammatory effects of an ethanol fraction from the Schisandra chinensis baillon hot water extract fermented using Lactobacilius paracasei subsp. tolerans. Food Science and Biotechnology. 2019;28(6):1759–1767. doi: 10.1007/s10068-019-00626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zofia N.-Ł., Aleksandra Z., Tomasz B., et al. Effect of fermentation time on antioxidant and anti-ageing properties of green coffee Kombucha Ferments. Molecules. 2020;25(22):5394. doi: 10.3390/molecules25225394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H., Lu Q., Liu R. Widely targeted metabolomics analysis reveals the effect of fermentation on the chemical composition of bee pollen. Food Chem. 2022;375 doi: 10.1016/j.foodchem.2021.131908. [DOI] [PubMed] [Google Scholar]
- 11.Liu, Minyan. Preparation of Total Glucoside of White Peony for Injection. (M.S.), Hebei Medical University, Available from: Cnki..
- 12.Chen S., Li X., Liu X., et al. Investigation of chemical composition, antioxidant activity, and the effects of Alfalfa flavonoids on growth performance. Oxid. Med. Cell. Longev. 2020;2020:1–11. doi: 10.1155/2020/8569237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Shi S., Yang N., et al. Comparative assessment of nutritional composition, polyphenol profile and antioxidative properties of wild edible ferns from northeastern China. Food Res. Int. 2023;163 doi: 10.1016/j.foodres.2022.112237. [DOI] [PubMed] [Google Scholar]
- 14.Cirlini M., Del Vecchio L., Leto L., et al. Sprouts of Moringa oleifera Lam.: Germination, polyphenol content and antioxidant activity. Molecules. 2022;27(24):8774. doi: 10.3390/molecules27248774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F., Huang G., Yang Z., et al. Antioxidant activity of Momordica charantia polysaccharide and its derivatives. Int. J. Biol. Macromol. 2019;138:673–680. doi: 10.1016/j.ijbiomac.2019.07.129. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y., Sun Y., Huang G. Preparation and antioxidant activities of important traditional plant polysaccharides. Int. J. Biol. Macromol. 2018;111:780–786. doi: 10.1016/j.ijbiomac.2018.01.086. [DOI] [PubMed] [Google Scholar]
- 17.Lucas-González R., Viuda-Martos M., Pérez Álvarez J.A., et al. Changes in bioaccessibility, polyphenol profile and antioxidant potential of flours obtained from persimmon fruit (Diospyros kaki) co-products during in vitro gastrointestinal digestion. Food Chem. 2018;256:252–258. doi: 10.1016/j.foodchem.2018.02.128. [DOI] [PubMed] [Google Scholar]
- 18.Li X. 2-Phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-Oxide (PTIO•) radical scavenging: a new and Simple antioxidant assay in vitro. J. Agric. Food Chem. 2017;65(30):6288–6297. doi: 10.1021/acs.jafc.7b02247. [DOI] [PubMed] [Google Scholar]
- 19.Yan B., Shen M., Fang J., et al. Advancement in the chemical analysis of Paeoniae Radix (Shaoyao) J. Pharmaceut. Biomed. Anal. 2018;160:276–288. doi: 10.1016/j.jpba.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Han Z., Zhang J., Zhang D., et al. Annual growth cycle observation, hybridization and forcing culture for improving the ornamental application of Paeonia lactiflora Pall. in the low-latitude regions. PLoS One. 2019;14(6) doi: 10.1371/journal.pone.0218164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Todorov S.D., Ivanova I.V., Popov I., et al. Bacillus spore-forming probiotics: benefits with concerns? Crit. Rev. Microbiol. 2021;48(4):513–530. doi: 10.1080/1040841X.2021.1983517. [DOI] [PubMed] [Google Scholar]
- 22.Nagy G., Vaz A.G., Szebenyi C., et al. CRISPR-Cas9-mediated disruption of the HMG-CoA reductase genes of Mucor circinelloides and subcellular localization of the encoded enzymes. Fungal Genet. Biol. 2019;129:30–39. doi: 10.1016/j.fgb.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Matsuyama T. Recent developments in terminator technology in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2019;128(6):655–661. doi: 10.1016/j.jbiosc.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Utpott M., Rodrigues E., Rios A.d.O., et al. Metabolomics: an analytical technique for food processing evaluation. Food Chem. 2022;366 doi: 10.1016/j.foodchem.2021.130685. [DOI] [PubMed] [Google Scholar]
- 25.Farhan M. Green Tea Catechins: Nature's Way of Preventing and treating Cancer. Int. J. Mol. Sci. 2022;23(18) doi: 10.3390/ijms231810713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabbir U., Rubab M., Daliri E.B.-M., et al. Curcumin, quercetin, Catechins and metabolic diseases: the role of Gut Microbiota. Nutrients. 2021;13(1):206. doi: 10.3390/nu13010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deepika Maurya P.K. Health benefits of quercetin in Age-related diseases. Molecules. 2022;27(8):2498. doi: 10.3390/molecules27082498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calderaro A., Patanè G.T., Tellone E., et al. The Neuroprotective Potentiality of flavonoids on Alzheimer's disease. Int. J. Mol. Sci. 2022;23(23) doi: 10.3390/ijms232314835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu X., Ma S., Gao Z., et al. One-step extraction and hydrolysis of flavonoid glycosides in Rape Bee pollen based on Soxhlet-Assisted Matrix Solid phase Dispersion. Phytochem. Anal. 2017;28(6):505–511. doi: 10.1002/pca.2699. [DOI] [PubMed] [Google Scholar]
- 30.Saisavoey T., Sangtanoo P., Chanchao C., et al. Identification of novel anti-inflammatory peptides from bee pollen (Apis mellifera) hydrolysate in lipopolysaccharide-stimulated RAW264.7 macrophages. J. Apicult. Res. 2020;60(2):280–289. [Google Scholar]
- 31.Theodosis-Nobelos P., Papagiouvannis G., Tziona P., et al. Antioxidant serine-(NSAID) Hybrids with anti-inflammatory and Hypolipidemic potency. Molecules. 2021;26(13):4060. doi: 10.3390/molecules26134060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X., Li D., Feng C., et al. Effects of hydrogen peroxide and l-tryptophan on antioxidative potential, apoptosis, and mammalian target of rapamycin signaling in bovine intestinal epithelial cells. J. Dairy Sci. 2022;105(12):10007–10019. doi: 10.3168/jds.2022-21869. [DOI] [PubMed] [Google Scholar]
- 33.Habte-Tsion H.M., Ren M., Liu B., et al. Threonine modulates immune response, antioxidant status and gene expressions of antioxidant enzymes and antioxidant-immune-cytokine-related signaling molecules in juvenile blunt snout bream (Megalobrama amblycephala) Fish Shellfish Immunol. 2016;51:189–199. doi: 10.1016/j.fsi.2015.11.033. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H., Li Z., Zhou S., et al. A fungal NRPS-PKS enzyme catalyses the formation of the flavonoid naringenin. Nat. Commun. 2022;13(1):6361. doi: 10.1038/s41467-022-34150-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.