Abstract
Genetic evolution of the simian immunodeficiency virus (SIV) envelope glycoprotein was evaluated in a group of six macaques (Macaca nemestrina) infected with the molecularly cloned, moderately pathogenic SIVsm62d. The extent of envelope evolution was subsequently evaluated within the context of the individual pattern of viremia and disease outcome. Two macaques in this cohort developed AIDS by 1.5 years postinoculation (progressors), whereas the remaining four macaques remained asymptomatic (nonprogressors). Compared with the nonprogressor macaques, the two progressor macaques exhibited higher persistent plasma viremia, higher homologous neutralizing antibody titers, and more extensive mutation and evolution in the V1 region of envelope. Although clearly distinct in each of these parameters from the progressors, the four nonprogressors exhibited more individual variability with respect to the extent of persistent viremia and genetic evolution of the V1 region of envelope. The extent of V1 envelope varied from no apparent V1 evolution in a macaque with good viral containment to extensive evolution in one macaque with persistent viremia. This study underscores the critical role of persistent replication in the genetic evolution of SIV.
Although human immunodeficiency virus, type 1 (HIV-1) is almost uniformly fatal, the rates of disease development and survival times vary widely between different individuals. Thus, there is a spectrum of disease progression from extremely rapid (1 to 2 years) to asymptomatic survival in excess of 10 years, with these extremes being observed at a low frequency (5, 8, 14, 39, 41, 49). The host and viral factors underlying variable disease progression have not been clearly defined and are likely to be complex. Clearly, the level at which plasma viremia stabilizes in the post-acute phase of infection has been shown to be an important prognostic indicator of disease progression in HIV infection (35, 36, 43, 52), and since down-modulation is coincident with development of cell-mediated immunity, a component of this effect may be due to a specific immune response (30).
Host factors which could potentially impact on disease progression include the level of immune activation at the time of infection, efficacy of the specific immune responses, antiviral factors elaborated by CD8+ T cells (11, 32, 33), and other undefined factors. Clear correlates of disease progression have not been discerned by comparison of the immune responses of progressors and nonprogressors. Both categories typically demonstrate strong, broadly neutralizing antibody responses (39, 45) as well as the presence of virus-specific cytotoxic T lymphocytes (as reviewed in reference 22). However, a subset of nonprogressors and exposed uninfected individuals have defects in the CCR5 gene (Δ32) which appear to render their cells less susceptible to infection with CCR5-utilizing viruses (reviewed in references 13 and 37). Viral factors such as virus load and the biologic phenotype of the virus infecting an individual could also affect disease progression. Many studies of the biologic evolution of HIV-1 demonstrated a phenotypic shift from early non-syncytium-inducing, macrophage-tropic virus strains early after seroconversion (50) to syncytium-inducing, T-cell-tropic strains later in the disease (2, 9, 20, 47, 48), possibly consistent with the emergence of more pathogenic variants. With the recent elucidation of the chemokine coreceptors required for HIV-1 entry, these phenotypes can largely be explained by a shift from CCR5-utilizing viruses to CXCR4-utilizing viruses late in disease (10, 16, 17–19). However, the relative pathogenicity of these two HIV-1 phenotypes is still unclear.
Genetic variation, selection, and evolution drive the biologic evolution of HIV-1 in vivo; each of these may be impacted by mutation rate, selective pressures, and replicative rate of the virus. A number of studies suggest that the evolution of the virus envelope glycoprotein differs depending upon the rate of disease progression (12, 15, 21, 53, 54). Studies of the rate of viral evolution of envelope and gag cytotoxic T lymphocytes epitopes in individuals with variable disease progression have demonstrated that the rate of evolution correlates inversely with the rate of disease progression, paradoxically, with the greatest evolution being observed in slow or nonprogressors; i.e., those individuals with low to moderate plasma viremia (12, 21, 54). The evolution of virus in rapid progressors or individuals with high viremia appears to be less extensive. These findings suggest a complex interplay between the rate of viral replication and selection pressure in driving viral evolution.
The simian immunodeficiency virus (SIV) macaque model has proven useful for the study of potential correlates of disease progression as well as the evolution of lentiviruses in vivo. The pathogenesis of SIV infection of macaques is remarkably similar to that of human AIDS (3, 4, 24, 26, 29, 31, 33, 34, 42, 46), albeit with a shorter period of clinical latency. As with HIV-1 infection, the level at which plasma viremia stabilizes following seroconversion is an important prognostic indicator (26, 51). The use of a molecularly cloned virus allows the investigator to study the roles of viral replication and immune pressure without confounding factors such as the complexity of the infecting quasispecies, selection at the time of infection, and the biologic phenotype of the infecting virus. The majority of studies of SIV evolution (1, 6, 7, 28, 43) have not taken into consideration such factors as the magnitude and specificity of the humoral immune response and the degree of viral replication as influencing the rate of viral evolution. Additionally, many of the molecularly cloned viruses examined were either more uniformly pathogenic (SIVmac239 [6, 29]) or minimally pathogenic (SIVsmH4 [23, 28]) (SIVmneC18 [44]), thus not allowing the investigation of evolution in animals exhibiting different disease courses. In the present study, we characterized the immune responses, sequential viral load, and genetic evolution of variable regions within the envelope glycoprotein in a group of six pigtailed macaques (Macaca nemestrina) inoculated with a molecularly cloned SIV, designated SIVsm62d. This virus contains gag-pol and vif genes of SIVsmH4 (a minimally pathogenic SIV [45]) and the vpx, tat, rev, env, and nef genes and 3′ long terminal repeat amplified in a single fragment directly from the splenic DNA of an SIVsm-infected pigtailed macaque (PT62) with AIDS as previously described (25) and is tropic for macaque CD4 lymphocytes and macrophages in vitro, with limited ability to infect any of a wide variety of human CD4+ T-cell lines.
Despite inoculation with a common molecularly cloned virus, the disease course was variable (25). Two animals (PT181 and PT182) exhibited depletion of peripheral CD4 lymphocyte numbers by 6 months postinoculation and progressed to AIDS with opportunistic infections by 1.5 years. The remaining four animals (PT185, PT187, PT188, and PT190) became infected as indicated by seroconversion and virus isolation from peripheral blood mononuclear cells (PBMC) but remained healthy throughout 3 years of observation. This spectrum of disease differs from that induced by more highly pathogenic molecularly cloned viruses, SIVmac239 (29) and SIVsmE543-3 (27), for which nonprogressors are rarely observed. The goal of this study was to evaluate evolution of the envelope variable regions as a potential predictor of progressive SIV infection and to develop a clearer understanding of the impact of viral replication rate in the selection of virus variants.
MATERIALS AND METHODS
Inoculation and evaluation of animals.
Six pigtailed macaques were inoculated intravenously with 1 ml of cell-free culture supernatant (approximately 1,000 50% tissue culture infective doses) from macaque PBMC infected with SIVsm62d, as described previously (25). Following inoculation, the animals were monitored sequentially by fluorescence-activated cell sorter analysis for lymphocyte subset changes (CD4, CD8, CD2, and CD20), virus isolation from PBMC, plasma viral load (see below), and SIV-specific antibody production by Western blot and neutralizing antibody assays (38). Virus isolation was conducted by stimulation of 5 × 106 PBMC with 10% interleukin-2 (IL-2) and phytohemagglutinin (PHA; 5 mg/ml) in RPMI 1640 medium supplemented with glutamine, Pen-Strep, and 10% fetal calf serum for 4 days, followed by cocultivation with an equal number of similarly stimulated PBMC from a normal macaque donor. Cultures were propagated for 6 weeks in RPMI 1640 supplemented as described above but without the addition of PHA and were fed twice weekly with a 50% media change. The culture supernatant was monitored weekly for the presence of reverse transcriptase activity. Total cellular DNA was isolated from sequential cryopreserved PBMC samples and tissues (axillary, inguinal, and mesenteric lymph nodes; spleen; thymus; and bone marrow aspirate), which were collected at autopsy as described previously (7).
Neutralizing antibody assays.
Neutralizing antibody titers to SIVsmH4 were assayed with a read-out of 50% inhibition of cell killing in CEMx174 cells as previously described (38) and thus were an assessment of the broadly reactive, homologous neutralization but not of neutralization escape. Neutralizing antibodies to SIVsm62d were assessed in phytohemagglutinin-stimulated pigtailed macaque PBMC as described previously (40), with minor modification. Briefly, cell-free virus was incubated with various dilutions of plasma samples in triplicate wells of 96-well culture plates for 1 h at 37°C. Following incubation, 30 μl was transferred to a second 96-well plate containing 3 × 105 PBMC in 150 μl of IL-2-containing growth medium. An additional six wells of cells received an equivalent amount of virus that had not been incubated with a plasma sample (virus control). Plates were incubated at 37°C for 4 h, and the medium was aspirated and replaced with 20 μl of fresh IL-2 growth medium. Medium was replaced an additional three times over the next 24 h to remove virus inoculum and anti-p27 antibodies in order to ensure later accurate quantitation of p27 production. Culture supernatants were collected daily for 13 days, mixed with 225 μl of 0.5% Triton X-100, and stored at 4°C for later p27 assays. Concentrations of p27 in virus control wells were quantified for each harvest day with a commercial p27 antigen capture assay as described by the supplier (Organon-Teknika/Azko, Durham, N.C.). Concentrations of p27 in the remaining wells were quantified when p27 production in the virus control wells was in a linear phase of increase and averaged 1,000 pg/ml. Neutralization titers are the highest plasma dilution at which p27 production was reduced by >90% compared to a corresponding dilution of the respective prebleed sample. All prebleed samples were negative for neutralization. Because residual anti-p27 antibody in test wells has the potential to interfere with the p27 assay, virus lysates were mixed from wells that were positive for neutralization with a known amount of SIVsm62d viral lysate, and no evidence was found that the test samples interfered with p27 quantitation.
Quantitative competitive PCR assay for plasma vRNA.
A system for HIV-1 DNA quantitation based upon an internally controlled PCR-based assay (QC-PCR) was adapted for use in the SIV model, as previously described (26). The primer sequences are as follows: for S-GAG03, 5′-CAGGGAAiiAGCAGATGAATTAG-3′ (nucleotide 1359); for S-GAG04, 5′-GTTTCACTTTCTCTTCTGCGTG-3′ (nucleotide 1873), where i represents inosine. For quantitation of viral RNA in plasma, a reverse transcriptase PCR version of the QC-PCR procedure was employed with an in vitro run-off transcript from pSGD83 as the internal control template. Plasma samples for analysis were collected with EDTA as the anticoagulant at 2, 6, 10, 18, 25, 42, 50, 58, 68, 77, and 85 weeks postchallenge and were stored in a −70°C freezer. Virions were pelleted by ultracentrifugation and lysed with sodium dodecyl sulfate-proteinase K, followed by serial organic extractions and precipitation with glycogen as a carrier. Replicate aliquots of the test RNA were subjected to reverse transcription with various known copy numbers of the in vitro transcript with random primers at 42°C for 30 min. The resulting cDNA was then amplified (45 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 1 min), and the products were quantitated as for DNA QC-PCR analysis. Results were normalized to the volume of plasma extracted and expressed as SIV RNA copies per milliliter of plasma. Interassay variation was less than 20% (coefficient of variation).
Single-stranded conformational polymorphism of PCR products.
To assess genetic variability within the V1 and V3 regions of the SIV envelope, single-stranded conformational polymorphism (SSCP) of PCR products amplified from either PBMC or tissue DNA was analyzed as described previously (7) with PCR with env primers 5′TGGGATGTCTTGGGAATCAGCTGCTTA (nucleotide 6588) and 5′CTTTTCTTGCTGAATTTGTGCTTCTTC (nucleotide 8596), followed by nested amplification with V1- or V3-specific primers and inclusion of 0.5 mCi of [α-32P]dCTP in the PCR mixture. Three microliters of the PCR product was diluted 1:1 in Sequenase stop buffer (U.S. Biochemicals), boiled for 5 min, and electrophoresed on a 10% glycerol-6% Hydrolink gel (AT Biochem, Malvern, Pa.) in 0.6× Tris-borate–EDTA running buffer. Primers used for SSCP are listed below, with nucleotide positions within the SIVsmH4 genome indicated in parentheses. The V1 primers were 5′-CTCACCCCACTATGTATAGCAATGAGA (6896) and 5′-AATTACAACCTATCATGGGCTCCTGTT (7098); the V3 primers were 5′-GATCAAGCTTAATAAGTATTATAATCTAAC (7493) and 5′-GATCCTCGAGTCTACAATTTGTCCACAT (7774).
Cloning and sequence analysis of viral DNA in sequential PBMC samples.
Envelope clones were obtained from PBMC or tissue samples following nested PCR amplification as previously described (7) with primers 5′TGGGATGTCTTGGGAATCAGCTGCTTA (nucleotide 6588) and 5′CTTTTCTTGCTGAATTTGTGCTTCTTC (nucleotide 8596). PCR-amplified products were digested with SphI and Csp45I and cloned into the plasmid vector pGEM-7Zf, which had been digested with the same restriction enzymes. The ligation mixtures were transformed into JM109, and all subsequent amplification steps were performed at 30°C. Ten to twenty colonies from each sample were identified by colony hybridization with a radiolabelled probe of the entire env gene, and hybridizing colonies were used in subsequent SSCP and sequencing reactions. Clones obtained from PBMC samples were sequenced manually by the dideoxy chain termination method (Sequenase), whereas clones obtained from tissues were sequenced with the automated ABI 373 sequencer. Nucleotide sequences and predicted amino acid sequences were analyzed with GeneWorks (Oxford Molecular) and Clustal V for multiple alignments.
RESULTS
The disease course in a group of six macaques infected with SIVsm62d was variable, with animals following one of two distinct clinical outcomes. Two macaques were defined as progressors based upon the development of AIDS-related disease, including peripheral CD4 lymphocyte depletion, opportunistic infections, or other AIDS-related syndromes, such as thrombocytopenia (PT181 and PT182). Four macaques which maintained normal CD4 lymphocyte subsets were considered nonprogressors or slow progressors (PT185, PT187, PT188, and PT190). Rapid progression, as defined by high escalating viremia with an ineffective humoral immune response and death from AIDS within 6 months of inoculation (27), was not observed in any of the SIVsm62d-infected animals.
Clinical and virologic characteristics of progressors and nonprogressors.
The progressor macaques exhibited declining peripheral CD4 lymphocyte subsets within 6 months of inoculation and chronic wasting within 1 to 1.5 years (Fig. 1). Both animals had other indirect signs of SIV infection, including anemia, thrombocytopenia, and lymphadenopathy (data not shown), although these signs predominated in PT182. At the time of autopsy, pathologic examination of tissues from PT181 revealed severe generalized depletion of all lymphoid tissues examined and disseminated granulomas due to Mycobacterium avium infection (25). Although peripheral CD4 lymphocytes were severely depleted, lymphoid depletion was more moderate in tissues of PT182; this macaque was severely anemic and thrombocytopenic at the time of autopsy (25). In situ hybridization (ISH) for SIV RNA revealed moderate numbers of SIV-expressing cells (five per high-power field) in lymphoid tissues of both progressor macaques, PT181 and PT182.
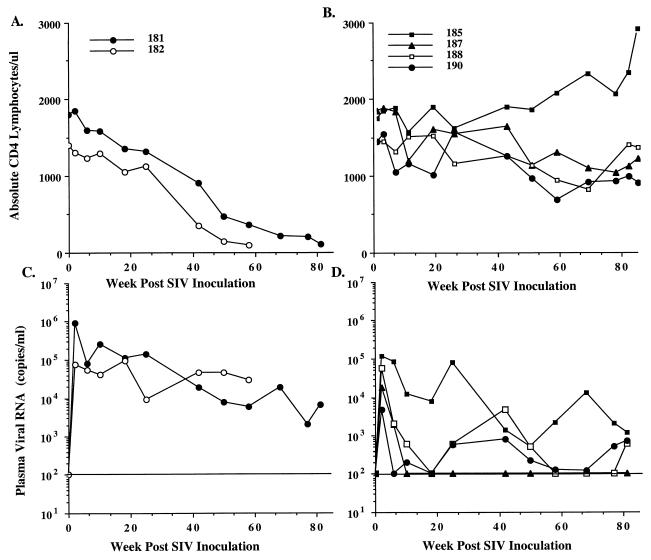
FIG. 1.
Sequential alterations in absolute circulating CD4 lymphocyte subsets and plasma viral load in SIVsm62d-infected macaques. Absolute peripheral CD4 lymphocytes in two SIVsm62d-infected progressor macaques (A) and four nonprogressors (B) and sequential viral RNA levels in plasma of two SIVsm62d-infected progressor (C) and four nonprogressor (D) macaques are shown. The horizontal line indicates the limits of quantitation of the assay for viral RNA in plasma.
The four nonprogressors remained clinically healthy throughout the period of observation (Fig. 1B) and maintained normal hematocrits, CD4 lymphocyte subsets, platelet counts, lymph node size, and weight gain (data not shown). At the time of euthanasia (approximately 3 years postchallenge), a complete necropsy of these animals revealed minimal pathologic changes. In particular, lymphoid tissues such as lymph nodes, spleen, and thymus were within normal limits, exhibiting neither lymphoid hyperplasia (a sign of early progression) nor lymphoid depletion. ISH of tissues collected from the nonprogressors failed to reveal SIV-expressing cells, and trapping of virus within germinal centers of lymphoid tissues was not observed (data not shown).
Viremia as a correlate of progression.
Although virus isolation from PBMC was generally successful early in the disease course, isolation from the nonprogressors became inconsistent after the first 4 months of infection (Table 1). In contrast, virus isolation from the progressors remained fairly consistent throughout the course of infection. Sequential plasma viral load was assessed, as shown in Fig. 1; all animals demonstrated primary plasma viremia at 2 weeks postinoculation. The level of plasma viremia ranged from 4,800 (PT190) to 910,000 (PT181) copies per ml. Both progressor macaques exhibited high primary viremia and subsequently maintained moderate plasma viral RNA levels into the post-acute phase of infection. Although plasma viremia in each of the progressor macaques remained at moderate levels for the first 6 months of infection, subsequent levels decreased to measurable but low levels in one animal (PT181); the decrease in plasma viremia was coincident with or preceded a precipitous decline in circulating CD4 lymphocytes and the onset of AIDS-like symptoms. Similar declining viremia in SIV-infected macaques in the terminal stages of AIDS associated with severe generalized lymphoid depletion and loss of susceptible target cells has been observed previously in SIVsm infection (25).
TABLE 1.
Virus isolation from PBMC of SIVsm62d-infected macaques
| Macaque no. | Virus isolation at indicated weeks after SIV inoculationa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 6 | 10 | 16 | 25 | 42 | 50 | 58 | 66 | 77 | 85 | |
| Progressors | |||||||||||
| PT181 | + | + | + | + | + | − | + | − | − | + | dead |
| PT182 | + | + | + | + | + | + | + | + | dead | dead | dead |
| Nonprogressors | |||||||||||
| PT185 | + | + | + | + | + | − | − | + | − | − | − |
| PT187 | + | + | − | − | − | − | − | − | − | − | + |
| PT188 | + | − | + | − | − | − | − | − | − | − | − |
| PT190 | + | + | − | − | − | − | − | − | − | + | − |
A plus sign indicates that virus was isolated from 5 × 106 PBMC within 6 weeks of coculture with macaque PBMC, whereas a minus sign indicates that virus was not isolated at this time point.
The nonprogressor group was more heterogeneous with respect to the pattern of plasma viremia than the progressors (Fig. 1). One animal exhibited rapidly declining primary plasma viremia to below detectable limits (PT187), and in two others, plasma viral RNA levels declined to consistently <500 copies per ml. However, one animal in this group (PT185) initially maintained plasma viral RNA levels more similar to those observed in the progressors. However, by 42 weeks postinfection, viral RNA levels had declined to 1,000 to 2,000 copies per ml without a concomitant decline in CD4 lymphocytes.
Neutralizing antibody responses.
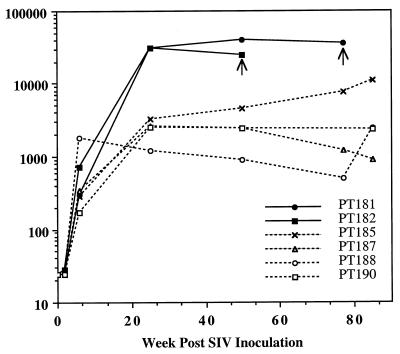
As a measure of functional, envelope-specific antibody, sequential neutralizing antibody titers against the related SIVsmH4 were evaluated in all six macaques (Fig. 2). SIVsmH4 has 96% identity in the predicted amino acid sequence of the envelope glycoprotein with SIVsm62d, and thus neutralizing titers should be a good indicator of homologous neutralizing antibody responses. Progressors developed neutralizing titers that were 10-fold higher than those of the nonprogressors. The nonprogressor PT185, which was an outlier in terms of plasma viral load, was also intermediate in antibody response between the progressors and nonprogressors, achieving a titer of 1:11,000. In general, the strength of the antibody response in the animals mirrored the degree of plasma viremia. Thus, PT187, in which plasma viremia was below detection limits for the majority of the observation period, exhibited declining neutralizing antibody titers. To confirm that neutralization activity against SIVsmH4 was a valid assessment of neutralizing antibody responses, plasma samples collected at peak titers (26 weeks) were evaluated for their ability to neutralize the inoculum, SIVsm62d. SIVsm62d proved to be highly sensitive to in vitro neutralization, unlike SIVsm543-3 (27). A trend in neutralization activity against SIVsm62d similar to that seen previously with SIVsmH4 was observed. Thus, both progressor macaques achieved titers of >1:625. The two nonprogressors (PT185 and PT190) with intermediate titers to smH4 also achieved titers of >1:625. The two nonprogressors with the lowest neutralizing antibody activity to smH4 exhibited lower neutralizing activity to SIVsm62d (1:125). Since endpoint titers were not achieved in this assay, we were unable to confirm that the progressors attained higher neutralization activity than the nonprogressors.
FIG. 2.
Reciprocal neutralizing antibody titers in SIVsm62d-infected macaques. Sequential SIV neutralizing titers are shown for the six SIVsm62d-infected macaques, with filled symbols and solid lines for the two progressors and dotted lines and open symbols for the nonprogressors. Arrows indicate time of death.
Evolution of envelope in PBMC of progressors and nonprogressors.
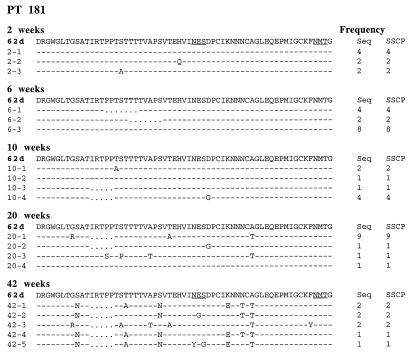
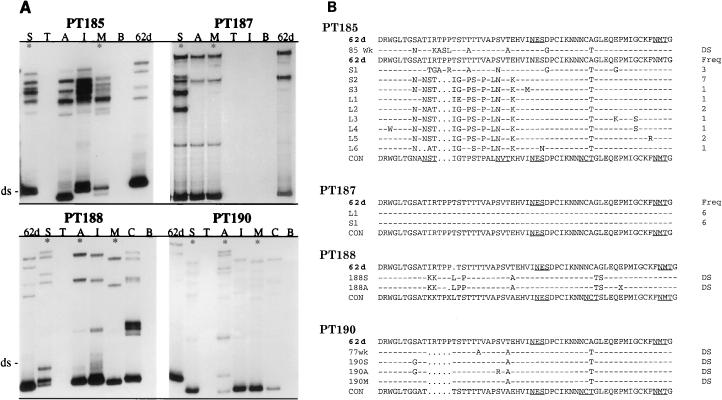
We used a combination of single-stranded conformational polymorphism (PCR-SSCP), sequence analysis of individual clones and direct sequence of PCR products to evaluate the extent of evolution within the most variable region of the SIV envelope glycoprotein, the V1 region (6, 28, 44). Initial analysis of the evolution of the V1 region within PBMC of one of the progressors demonstrated that deletions were observed as early as 6 weeks postinoculation (Fig. 3), with substitutions which introduced new N-linked glycosylation sites observed later in the course of infection (20 weeks). Similar analysis of viral evolution in the nonprogressors was not feasible due to low PBMC viral load that was evident in SSCP analysis (data not shown). Therefore, we subsequently focused on evaluation of V1 evolution in tissues. PCR-SSCP analysis of the V1 region confirmed widespread distribution of proviral DNA in various lymphoid tissues with the exception of thymus and bone marrow of each of the nonprogressor macaques (Fig. 4A). The predominant virus population in tissues of all but one of the macaques (PT187) was distinct from the inoculum, based on mobility of the double-stranded and single-stranded products.
FIG. 3.
Alignment of predicted amino acid sequences of the V1 region of PCR products cloned from PBMC of PT181 at sequential time points during infection. The sequence of SIVsm62d is shown at the top of each time point. Identity at a residue is indicated with a dash, dots indicate a deletion relative to SIVsm62d, and amino acid substitutions are shown with the single-letter amino acid code. At the right is shown the frequency of the number of clones of each unique sequence type as determined by sequence and SSCP analysis.
FIG. 4.
Analysis of genetic variation in the V1 region of envelopes cloned from tissues of nonprogressors. (A) SSCP analysis of the V1 region of tissues of SIVsm62d-infected nonprogressor macaques (185, 187, 188, and 190). Asterisks indicate tissues used for cloning and/or sequence analysis. 62d, SIVsm62d; ds, double stranded; S, spleen; T, thymus; A, axillary lymph node; I, inguinal lymph node; M, mesenteric lymph node; C, ileocecal junction; B, bone marrow. Migration of the double-stranded band indicates deletions or insertions; differences in mobility of the single-stranded bands indicate nucleotide substitutions; single-stranded bands in addition to the two expected for cloned virus indicate quasispecies within the sample (such as those observed for PT182 and PT185). (B) Alignment of predicted amino acid sequences of the V1 region encoded by env from PCR products cloned from tissues (spleen and lymph node) of PT185 and PT187 and direct sequence analysis (DS) of PCR products amplified from spleen and lymph nodes of PT188 and PT190. The sequence of SIVsm62d is shown at the top, and the consensus sequence of the virus in the macaque is shown at the bottom of each alignment (CON). Identity at a residue is indicated with a dash, dots indicate a deletion relative to SIVsm62d, amino acid substitutions are shown with the single-letter amino acid code, and an X substitution indicates polymorphism in the nucleotide sequence at this point. At the right is shown the frequency of the number of clones of each unique sequence type as determined by sequence and SSCP analysis.
Since relative changes in SSCP mobility are not predictive of actual nucleotide divergence, individual V1 clones were cloned from tissues of the two progressors (PT181 and PT182) and two nonprogressors (PT185 and PT187), and PCR products amplified from tissues of the other two nonprogressors (PT188 and PT190) were directly sequenced. As summarized in Table 2, almost all of the nucleotide changes were nonsynonymous, and therefore most resulted in changes in the predicted amino acid sequence. However, in some instances, two or three nucleotide positions in the codon were altered, and therefore the number of nucleotide substitutions was somewhat higher than the number of respective amino acid substitutions. Analysis of the respective predicted protein sequences of V1 from various macaques revealed significant evolution of virus in five of the macaques, three of the nonprogressors (Fig. 4B), and the progressors (Fig. 5). The extent of V1 evolution in the two progressor macaques was similar and fairly extensive. In contrast, V1 evolution within the nonprogressors was variable (Fig. 4). Thus, at one end of the spectrum, the V1 region of env within tissues of PT187 was essentially unchanged from the input virus. In contrast, the V1 region of virus within tissues of PT185 was distinct from the input virus, and the extent of evolution was similar in magnitude to that observed in the progressors macaques. An intermediate degree of evolution was observed for two nonprogressor macaques, PT188 and PT190; the predominant viral genotype was distinct from that of the inoculum but was considerably less divergent than that observed in the progressor macaques. Changes relative to the original inoculating virus included deletions as well as substitutions. As shown in Fig. 4 and 5, some substitutions resulted in the introduction of potential N-linked glycosylation sites (three more for PT181, one for PT182, three for PT185, and one for PT188 and PT190).
TABLE 2.
Relationship between mean post-acute plasma viremia and genetic evolution of envelope V1 region
| Macaque no. | Wk postinfection | No. of amino acid
|
No. of nucleotide substitutions | Mean level of plasma viral RNA | Mean no. of amino acid substitutions/wka | |
|---|---|---|---|---|---|---|
| Substitutions | Deletions | |||||
| PT181 | 76 | 13 | 5 | 13 | 64,500 | 0.171 |
| PT182 | 51 | 12 | 5 | 13 | 46,000 | 0.235 |
| PT185 | 136 | 12 | 5 | 18 | 19,500 | 0.095 |
| PT187 | 132 | 0 | 0 | 0 | 900 | 0.000 |
| PT188 | 136 | 8 | 1 | 8 | 300 | 0.058 |
| PT190 | 136 | 4 | 5 | 4 | 200 | 0.029 |
The mean number of substitutions per week was calculated by dividing the maximum number of substitutions by the week postinfection when the sequence was evaluated.
FIG. 5.
Alignment of predicted amino acid sequences of the V1 region encoded by env from PCR products cloned from tissues (spleen and lymph node) of PT181 and PT182, which were collected at the time of autopsy. The sequence of SIVsm62d (62d) is shown at the top, and the consensus sequence of the virus in the macaque is shown at the bottom of each alignment (CON). Potential N-linked glycosylation sites are underlined in the 62d sequence and consensus. Identity at a residue is indicated with a dash, dots indicate a deletion relative to SIVsm62d, and amino acid substitutions are shown with the single-letter amino acid code. At the right is shown the frequency of the number of clones of each unique sequence type as determined by sequence and SSCP analysis.
The rate of viral evolution appeared to correlate with the magnitude of persistent viremia. The evolution of viral sequences within the nonprogressors was generally slower than in the progressors but varied significantly between animals. Table 2 shows the relationship between the extent of plasma viremia (mean post-acute plasma viral RNA) and the rate of V1 evolution as assessed by determining the number of amino acid changes per week. Evolution of V1 was greatest in the two progressors and then decreased in rank order, with decreasing mean plasma viremia. The evolution of the V1 region was most pronounced in PT185, an animal which exhibited persistent moderate plasma viremia throughout the course of infection. The V1 region evolved to a lesser extent and/or rate in animals with intermittently detectable post-acute plasma viremia (PT188 and PT190). The least evolution was observed in clones obtained from the macaque for which viral containment was most restricted and plasma viral RNA was undetectable after primary viremia (PT187). However, since the strength of the neutralizing antibody response covaried with the level of plasma viremia, both must be considered potential factors controlling evolution.
DISCUSSION
Persistent high virus load appeared to be a major correlate of progressive disease in SIV infection, as has been observed in previous studies with uncloned SIV isolates (26, 51). Since the SIVsm62d is moderately pathogenic compared to the highly pathogenic SIVmac239, the spectrum of disease course in this cohort was shifted toward a predominance of animals with slowly progressive or nonprogressive disease. None of the SIVsm62d-infected macaques exhibited rapid progression. The levels of peak viremia in this cohort are also in agreement with this assessment of the relative pathogenicity, since peak levels during acute viremia were significantly lower (103 to 106 copies/ml) than levels observed in infection with highly pathogenic SIV isolates (106 to 108 copies/ml) (26, 51).
In terms of genetics, this study underscores previous observations that the evolution of virus in an individual can be highly variable and clearly does not proceed at a linear rate throughout the time course of infection. Additionally, this study begins to delineate some of the factors responsible for variation in viral evolution among individuals. Factors such as the persistence of viral replication and the magnitude of the humoral immune response clearly played a pivotal role in the process of in vivo evolution. In the present study, the strength of the humoral immune response correlated directly with the amount of ongoing viral replication, making it difficult to assess the relative contributions of these two parameters to in vivo evolution. The assessment of the role of neutralizing antibody in this study was further complicated by the use of a highly homologous virus, rather than the inoculum or later virus isolates from the infected macaques, as the target for neutralization assays. Thus, we were unable to evaluate whether these macaques were undergoing sequential evolution of neutralization-resistant variants. Overall, both antibody selection and ongoing viral replication are likely to be essential for promoting viral evolution of SIV. The evolution of new N-linked glycosylation sites in the V1 region is consistent with attempts by the virus to evade neutralizing antibody responses. Interestingly, we observed viral evolution even in the nonprogressor macaques which had intermittent, extremely low but detectable plasma viremia (<1,000 copies/ml). The only macaque for which evolution was not observed was one in which plasma viremia was undetectable throughout most of the course of infection.
The present macaque study suggests a correlation between the rate of evolution of the SIV envelope and the level of persistent plasma viremia in the infected macaque. Since the persistence of viremia correlated with disease course, viral evolution was greatest in the progressor macaques and least in the nonprogressors. The findings of the present study differ from previous reports of HIV-1 evolution. In HIV-infected individuals, viral load and evolution appear to be inversely correlated (15, 21, 54); rapid progressors with high viremia exhibited less viral evolution and less viral diversity than individuals with slower progression. The differences between this macaque study and those with HIV-infected humans could be due to a number of factors, including the clonality or heterogeneity of the virus inoculum, differences in the biologic phenotype of HIV-1 infecting different individuals, and the relative levels of viral replication in the individuals chosen for study. Studies of HIV-infected individuals have concentrated mainly upon rapid progressors and slow progressors. In contrast, rapid progressors were not evaluated in the present macaque study, and the levels of viremia in the macaque were significantly lower than in the HIV-infected individuals studied. For example, in the pediatric studies, plasma viremia in the children with slowly progressive disease was characteristically in excess of 104/ml, whereas plasma viremia in our SIV-infected macaque nonprogressors was at least an order of magnitude lower (<103/ml). Our nonprogressor macaques were more comparable in levels of viremia to HIV-infected individuals undergoing highly active antiretroviral therapy. Indeed, viral evolution appears to be minimal in these individuals, entirely consistent with the results of our study. The moderately pathogenic nature of the molecularly cloned SIV used in the study precludes evaluation of in vivo evolution during rapid progression, which will be an obvious area of future interest.
In conclusion, the SIV model offers the luxury of a known molecularly cloned inoculum, dose, and timing of inoculation that would not be possible in studying cohorts of HIV-infected individuals. Such a study allows us the opportunity to evaluate the relative contribution of selective pressures, such as antibody and the rate of viral replication, to the in vivo evolution of SIV in macaques and the subsequent analogy of HIV-1 in humans.
ACKNOWLEDGMENTS
We thank Robert Chanock for his support in performing this study, Russell Byrum for conducting the animal studies, and Malcolm Martin for helpful comments in the writing of the manuscript.
REFERENCES
- 1.Anderson M, Hauer D, Sharma D P, Joag S V, Narayan O, Zink M C, Clements J E. Analysis of envelope changes acquired by SIVmac239 during neuroadaptation in rhesus macaques. Virology. 1993;195:616–626. doi: 10.1006/viro.1993.1413. [DOI] [PubMed] [Google Scholar]
- 2.Asjo B, Albert J, Karlsson A, Mordfeldt-Manson L, Bibberfeld G, Lidman K, Fenyo E M. Replicative properties of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 3.Baskin G, Martin L N, Murphey-Corb M, Hu F-S, Kuebler D, Davison B. Distribution of SIV in lymph nodes of serially sacrificed rhesus monkeys. AIDS Res Hum Retroviruses. 1995;11:273–285. doi: 10.1089/aid.1995.11.273. [DOI] [PubMed] [Google Scholar]
- 4.Baskin G B, Murphey-Corb M, Watson E A, Martin L N. Necropsy findings in rhesus monkeys experimentally infected with cultured simian immunodeficiency virus (SIV/Delta) Vet Pathol. 1989;25:456–467. doi: 10.1177/030098588802500609. [DOI] [PubMed] [Google Scholar]
- 5.Buchbinder S P, Katz M H, Hessol N A, O’Malley P M, Holmberg S D. Long-term HIV-1 infection without immunological progression. AIDS. 1994;8:1123–1128. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 6.Burns D, Desrosiers R. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell B, Hirsch V M. Extensive envelope heterogeneity of simian immunodeficiency virus in tissues from infected macaques. J Virol. 1994;68:3129–3137. doi: 10.1128/jvi.68.5.3129-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y, Qin L, Zhang L, Safrit J, Ho D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 9.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 10.Choe H, Farazan M, Sun Y, Sullivan N, Rollins B, Panath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 11.Cocci P F, DeVico A, Garazino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α and MIP-1β as the major immunosuppressive factors produced by CD8+ T cells. Science. 1995;270:483–489. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 12.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 13.Cohen O J, Kinter A, Fauci A S. Host factors in the pathogenesis of HIV disease. Immunol Rev. 1997;159:31–67. doi: 10.1111/j.1600-065x.1997.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 14.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delwart E, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 18.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry co-factor: functional cDNA cloning of a seven transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 20.Fenyo E M, Norrby E. Biological variation of human immunodeficiency viruses and evolution of the late pathogenic infection in vivo. In: Koff W, Wong-Staal F, Kennedy R, editors. AIDS research reviews. 1st ed. New York, N.Y: Marcel Dekker, Inc.; 1991. pp. 149–164. [Google Scholar]
- 21.Ganeshan S, Dickover R E, Korber B T M, Bryson Y J, Wolinsky S M. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–667. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goulder P, Price D, Nowak M, Rowland-Jones S, Philips R, McMichael A. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol Rev. 1997;159:17–29. doi: 10.1111/j.1600-065x.1997.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch V, Olmstead R, Murphey-Corb M, Purcell R, Johnson P. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch V M, Johnson P R. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch V M, Martin J E, Dapolito G, Elkins W R, London W T, Goldstein S, Johnson P R. Spontaneous substitutions in the vicinity of the V3 analog affect cell tropism and pathogenicity of simian immunodeficiency virus. J Virol. 1994;68:2649–2661. doi: 10.1128/jvi.68.4.2649-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch V M, Adger-Johnson D, Campbell B, Goldstein S, Brown C R, Elkins W R, Montefiori D C. A pathogenic molecularly cloned simian immunodeficiency virus SIVsmE543-3 shares characteristics of primary human immunodeficiency virus isolates. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson P R, Hamm T E, Goldstein S, Kitov S, Hirsch V M. The genetic fate of molecularly cloned simian immunodeficiency virus in experimentally infected macaques. Virology. 1991;185:217–228. doi: 10.1016/0042-6822(91)90769-8. [DOI] [PubMed] [Google Scholar]
- 29.Kestler H, Kodama T, Ringler D, Marthas M, Pederson N, Lackner A, Reiger D, Seghal P, Daniel M, King N, Desrosiers R C. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 30.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal associations of cellular immune responses with the initial control of viremia in primary HIV-1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letvin N, King N. Immunologic and pathologic manifestations of the infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Acquired Immune Defic Syndr. 1990;3:1023–1040. [PubMed] [Google Scholar]
- 32.Levy J A, Makewicz C E, Barker E. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ cells. Immunol Today. 1996;17:217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- 33.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClure H M, Anderson D C, Fultz P N, Ansari A A, Lockwood E, Brodie A. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet Immunol Immunopathol. 1989;21:13–24. doi: 10.1016/0165-2427(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 35.Mellors J W, Kingsley L A, Rinaldo C R, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 36.Mellors J W, Rinaldo C R, Christopherson C, Sninsky J, Greenfield L, Kwok S. Prognosis of HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 37.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Shepard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 38.Montefiori D C, Robinson W E, Jr, Schuffman S S, Mitchell W M. Evaluation of antiviral drugs and neutralizing antibodies aginst human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26:231–235. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montefiori D C, Panteleo G, Fink L M, Zhou J T, Bilska M, Miralles G D, Fauci A S. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J Infect Dis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- 40.Montefiori D C, Baba T W, Li A, Bilska M, Ruprecht R M. Neutralizing and infection-enhancing antibody responses do not correlate with the differential pathogenicity of SIVmac239Δ3 in adult and infant rhesus monkeys. J Immunol. 1996;157:5528–5535. [PubMed] [Google Scholar]
- 41.Munoz A, Kirby A J, He Y D, Margolick J B, Visscher B R, Rinaldo C R, Kaslow R A, Phair J P. Long-term survivors with HIV-1 infection: incubation period and longitudinal patterns of CD4+ lymphocytes. J Acquired Immune Defic Syndr. 1995;8:496–505. doi: 10.1097/00042560-199504120-00010. [DOI] [PubMed] [Google Scholar]
- 42.Nowak M A, Lloyd A, Vasquez G, Wiltrout T A, Fischofberger N, Williams J, Kinter A, Fauci A S, Hirsch V M, Lifson J D. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien T, Blattner W A, Waters D, Eyster E, Hilgartner M W, Cohen A R, Luban N, Hatzakis A, Aledort L M, Rosenberg P S, Miley W J, Kroner B L, Goedart J J, et al. Serum HIV-1 RNA levels and time to development of AIDS in the multicenter hemophilia cohort study. JAMA. 1996;276:105–110. [PubMed] [Google Scholar]
- 44.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilgrim A K, Panteleo G, Cohen O J, Fink L M, Zhou J Y, Zhou J T, Bolegnesis D P, Fauci A S, Montefiori D C. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term nonprogressive infection. J Infect Dis. 1997;176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- 46.Reimann K, Tenner-Racz K, Racz P, Montefiori D, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2368–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richman D D, Bozzette S A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 48.Schuitemaker H, Koot M, Kootstra N A, Wouter Dercksen M, de Goede R E Y, van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheppard H W, Lang W, Ascher M S, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS. 1993;7:1159–1166. [PubMed] [Google Scholar]
- 50.Van’t Wout A B, Koostra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Countinho R A, Miedema F, Schuitemaker H. Macrophage tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S-L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G W. Viral dynamics in human immunodeficiency virus, type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 53.Wolf T F W, deJong J J, van der Berg H, Tijnagel J M G H, Krone W J A, Goudsmit J. Evolution of sequences encoding the principal neutralizing epitope of HIV-1 is host-dependent, rapid and continuous. Proc Natl Acad Sci USA. 1990;87:9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J F, Koup R A. Adaptive evolution of human immunodeficiency virus type 1 during the natural course of infection. Science. 1996;255:1134–1137. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]