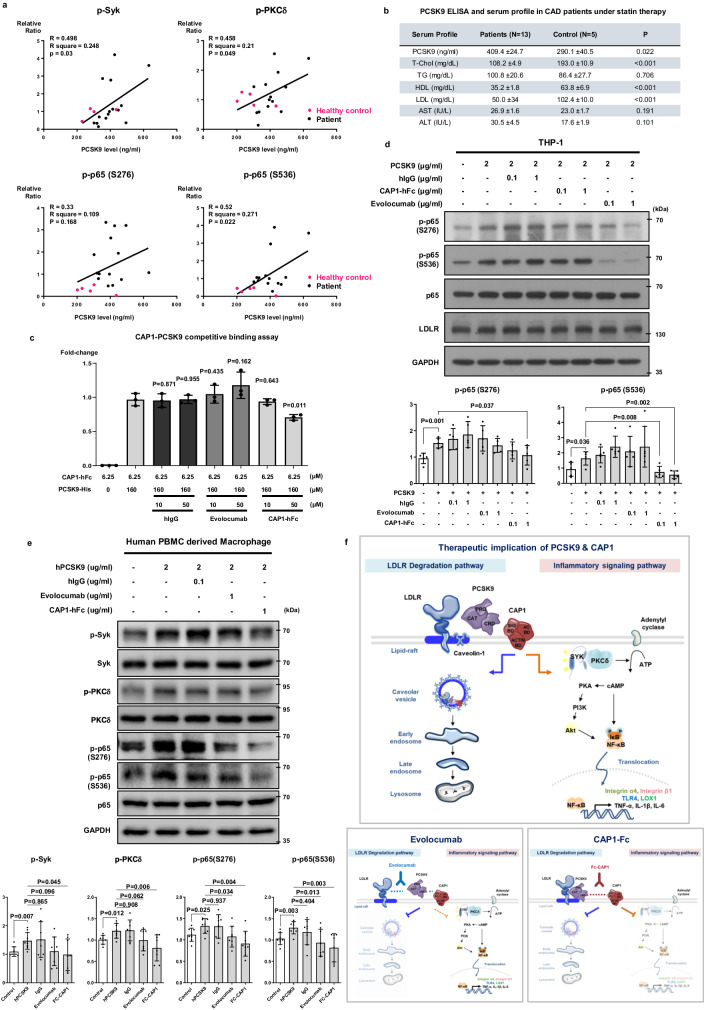
Fig. 10. PCSK9 levels in the serum of patients with coronary artery disease (CAD) correlated with Syk, PKC, and NF-κB phosphorylation, and PCSK9-mediated phosphorylation was blocked by CAP1-hFc.
a Simple linear regression analysis of Pearson correlation by distance. This figure illustrates the relationship between serum PCSK9 concentration and the quantified levels of phosphorylated proteins in a single individual. Serum PCSK9 concentration displayed a positive correlation with the phosphorylation of Syk, PKCδ, p65(S276), and p65(S536) in human peripheral blood mononuclear cells (PBMCs). The black line represents an asymptotic regression line fitted to the raw data (N = 19). b Average serum PCSK9 concentration and lipid profile results from 13 hyperlipidemia patients and five healthy donors. T-Chol total cholesterol, TG triglyceride, HDL high-density lipoprotein, LDL low-density lipoprotein, AST aspartate aminotransferase, ALT alanine aminotransferase. c Competitive ELISA binding assay showed that PCSK9-His bound to CAP1, and its interaction was competitively inhibited by CAP1-hFc, whereas evolocumab did not hinder the PCSK9-CAP1 interaction (N = 3). d Immunoblot analysis of NF-κB p65 signal activation after treatment with rhPCSK9 (2 µg/mL) along with human IgG, evolocumab, or CAP1-hFc in THP-1 cells revealed that the phosphorylation of p65 induced by PCSK9 was notably reduced by CAP1-hFc (N = 5). e Immunoblot analysis demonstrated that PCSK9-induced phosphorylation of Syk, PKCδ, and NF-κB p65 in human PBMC-derived macrophages was attenuated by CAP1-hFc. After 1-h treatment of rhPCSK9 with human IgG, evolocumab, or CAP1-hFc, the PCSK9-induced phosphorylation of Syk, PKCδ, and NF-κB p65 was significantly decreased by CAP1-hFc (N = 8). f Schematic diagram depicting CAP1 as the binding partner of PCSK9, which mediates not only caveolae-dependent endocytosis and lysosomal degradation of LDLR, but also recruits Syk and PKCδ and modulates PCSK9-mediated inflammatory signal transduction. CAP1-hFc inhibits the binding of CAP1 and PCSK9, which sequentially block LDLR degradation and the inflammatory signaling pathway, whereas the PCSK9 inhibitor evolocumab can only block the LDLR degradation pathway. The differences between the groups were compared using the unpaired t-test (two-tailed). All experiments are independently performed and all data are presented as mean values ± SD.