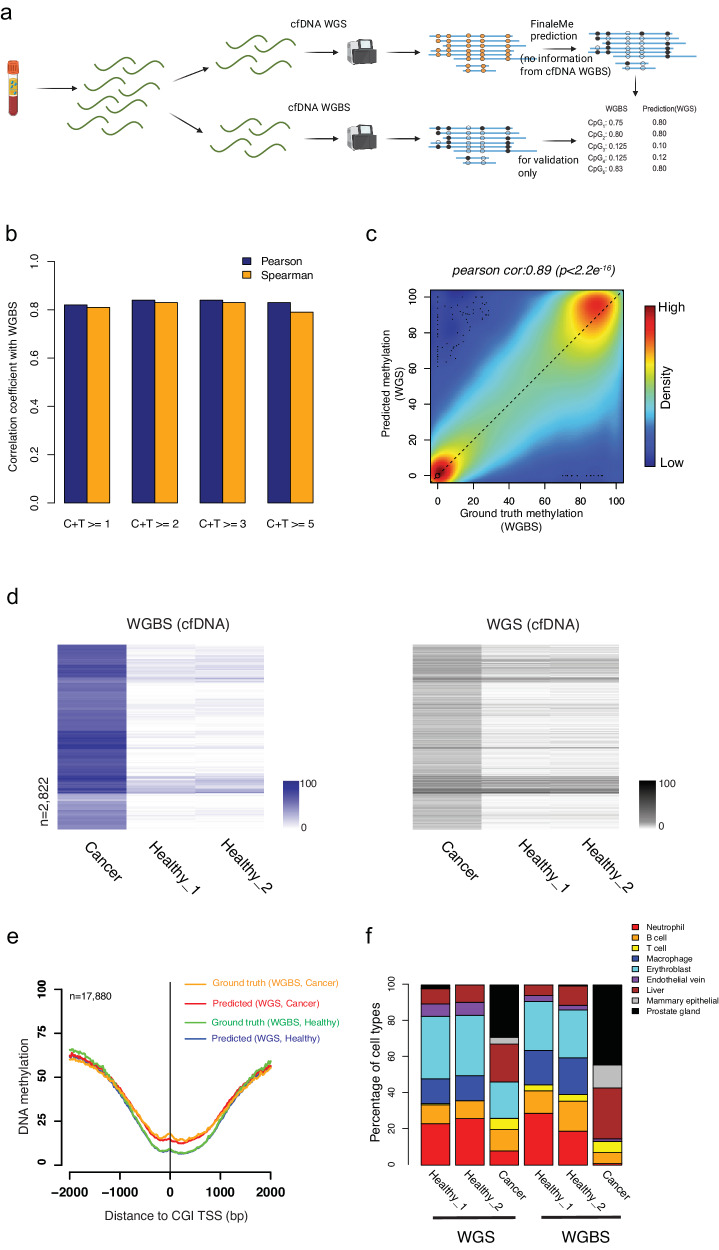
Fig. 3. Inferring DNA methylation from high-coverage whole-genome sequencing.
a Workflow to benchmark the model performance. Created with BioRender.com. b Pearson and Spearman correlation of DNA methylation at single CpGs with different coverages at CpG island and CpG island shore regions between matched cfDNA WGBS and WGS. Blue bars represent the Pearson correlation coefficient. Orange bars represent the Spearman correlation coefficient. c Scatterplot of DNA methylation level within 1 kb non-overlapped bins (n = 116,133) at CpG island and CpG island shore regions between matched cfDNA WGBS and WGS. The correlation coefficient and p-value is calculated by two sided Pearson correlation test in cor.test function in R. d Heatmap of measured (left panel, cfDNA WGBS, purple) and predicted (right panel, matched cfDNA WGS, black) DNA methylation level at hypermethylated differentially methylated windows (1 kb) characterized in CGI and CGI shore regions (n = 2822). The row orders in both WGBS and WGS datasets were based on the clustering of DNA methylation levels in WGBS only. e Average ground truth (WGBS) and predicted (WGS) DNA methylation level at CpG island promoter regions (n = 17,880) from cancer and healthy individuals. Orange line represents the ground truth from WGBS in the cancer patient. Red line represents the predicted value from WGS in the cancer patient. Green line represents the ground truth from WGBS in the healthy individual. Blue line represents the predicted value from WGS in the healthy individual. f The fraction of cell types that contributed to cfDNA was estimated by matched WGS and WGBS. Red: Neutrophil. Orange: B cell. Yellow: T cell. Blue: Macrophage. Cyan: Erythroblast. Purple: Endothelia vein. Brown: Liver. Gray: Mammary epithelia. Black: Prostate gland. Source data are provided as a Source Data file.