Figure 4.
Associations between gut microbiome alteration and sexual practices in MSM
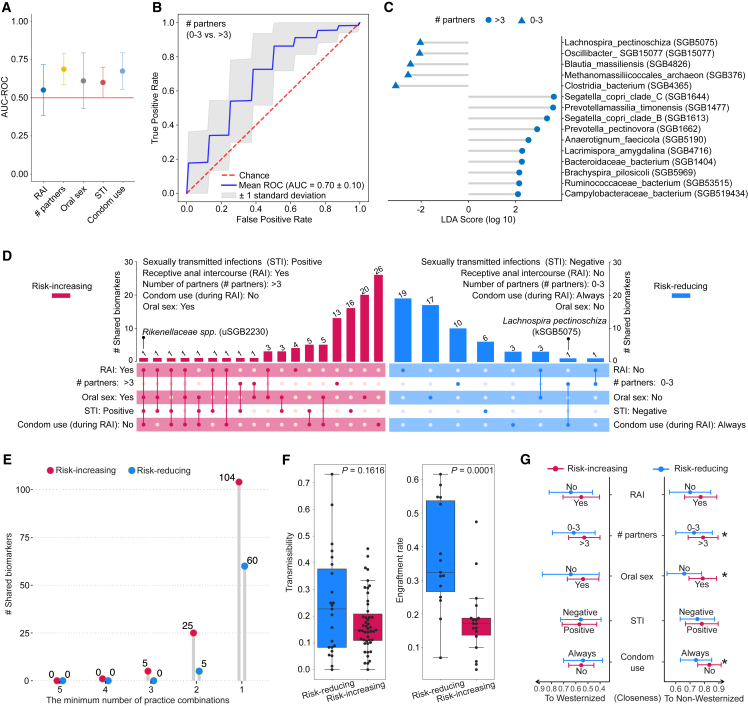
(A) Association analysis between the whole microbiome composition and sexual practices assessed by machine learning prediction. The approach was based on a 20-repeated 3-fold-stratified cross-validation using random forest approach, given species-level relative abundances as input.
(B) ROC (receiver operating characteristic) curve of the learning model based on the species-level relative abundances of individuals corresponding to having 0–3 and >3 partners (in the last 12 months) using a random-forest-based approach.
(C) Species-level taxonomic biomarkers identified using LEfSe58 to associate with behaviors having >3 (and 0–3) sexual partners. Only taxonomically known species are displayed (refer to Table S4 for a complete list).
(D) UpSet plot showing overlaps of taxonomic biomarkers identified using LEfSe for sexual practices classified as risk increasing and reducing. Numbers above bars indicate the number of biomarkers, solid dots denote sexual practices, biomarkers shared by multiple practices are denoted by connected dots, and biomarkers exclusively associated with a single sexual practice are denoted by single dots.
(E) The number of taxonomic biomarkers shared by the minimum number of practice combinations in risk-increasing and -reducing categories.
(F) Boxplots showing the transmissibility (left) and engraftment rate (right) of taxonomic biomarkers associated with risk-increasing and -reducing practices.
(G) Closeness of the gut microbiome to Westernized and non-Westernized populations with respect to sexual practices and STIs. Circles show the AUC-ROC mean and error bars represent the standard deviation. Asterisk denotes the statistical significance (∗p < 0.05) by Wilcoxon rank-sum test.