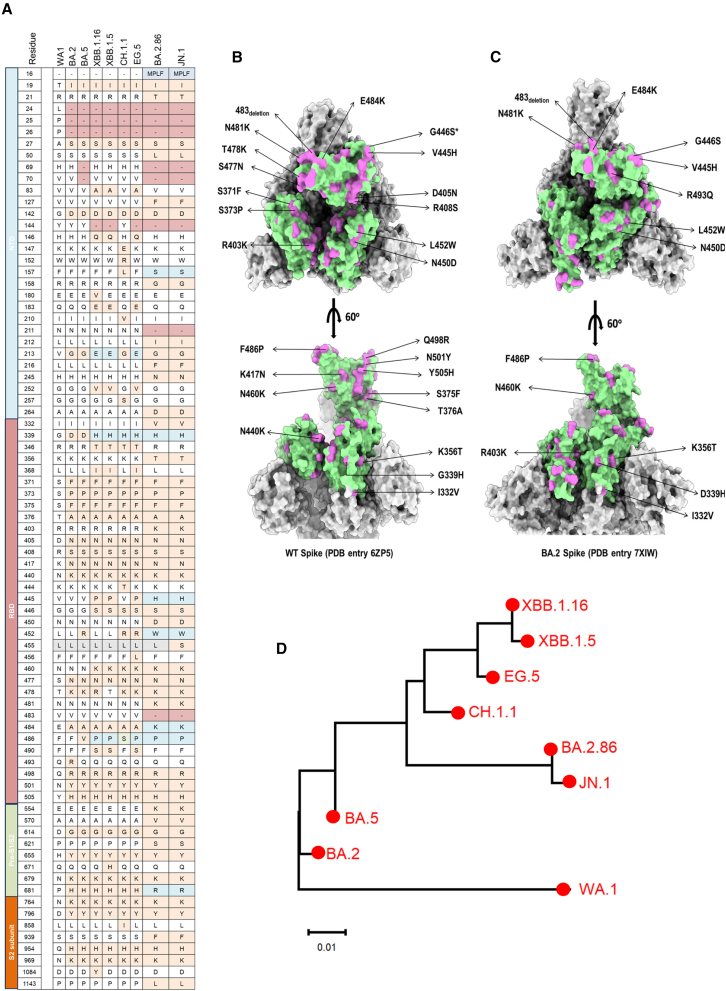
Figure 1.
Sequence comparisons between BA.2.86, JN.1, and other representative Omicron subvariants
(A) Mutations in S proteins of BA.2, BA.5, XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 relative to original SARS-CoV-2 wild-type strain (WA1).
(B and C) Amino acid mutation sites in RBD of BA.2.86 compared with wild-type SARS-CoV-2 and BA.2. S trimers of BA.2 (PDB: 7XIW) and wild type (PDB: 6ZP5) are shown in a surface representation, colored in gray. The RBD region is colored in pale green. The mutations of BA.2.86 in the RBD, in comparison to BA.2 and the wild-type strain, are indicated and colored in orchid.
(D) Phylogenetic tree based on the RBD amino acid sequences shows the genetic distance between the SARS-CoV-2 wild-type strain (WA1) and its variants and subvariants, BA.2, BA.5, XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1. The GeneBank accession numbers for their S protein amino acid sequences are GeneBank: YP_009724390.1 (WA1); GeneBank: WNO30888.1 (BA.2); GeneBank: WNH55069.1 (BA.5); GeneBank: WPU07054.1 (XBB.1.5); GeneBank: WNN93469.1 (XBB.1.16); GeneBank: WLS36136.1 (CH.1.1); GeneBank: WNK91259.1 (EG.5); GeneBank: WNK92404.1 (BA.2.86); and GeneBank: WQN42696.1 (JN.1).