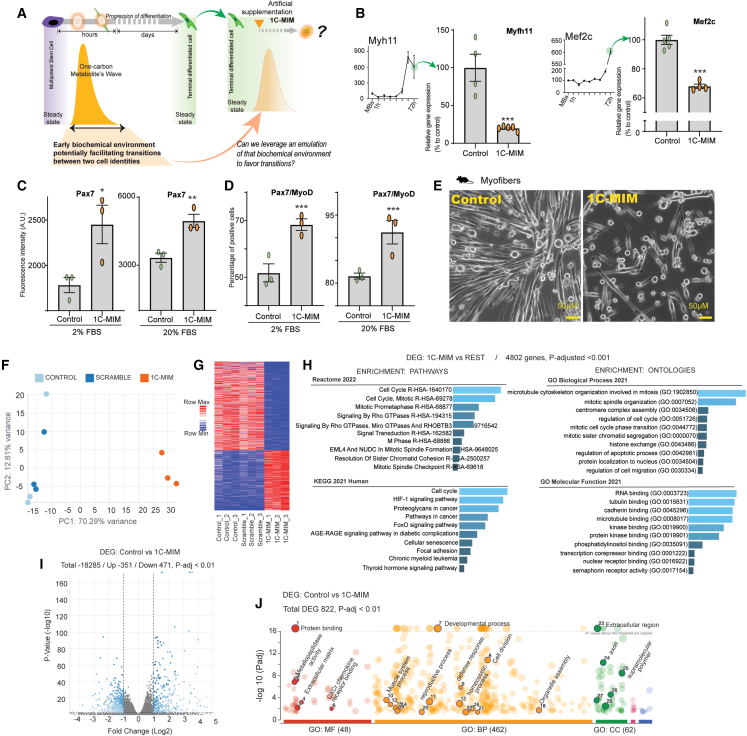
Figure 2.
Mirroring the biochemical transitional niche evokes the reduction of differentiation marks on mature cells
(A) Schematic representation of the supplementation of metabolites to partially imitate a 1C (one-carbon) wave with 1C-MIM (metabolite induction medium) over a terminally differentiated steady state. Note that the 1C-MIM represents an artificial intervention to be added in mature cells that do not have a 1C wave.
(B) Effects on the supplementation of 1C-MIM over the differentiation markers of myofibers Myfh11 (on the left) and Mef2c (on the right). Expression evaluated by RT-PCR. Bars represent the gene expression levels on differentiated control myofibers or those exposed for 3 days to the supplementation. Top left insets show the levels observed during differentiation. The gene expression of genes of interest was normalized with the geometric mean of at least two housekeeping genes (from Actb, Gapdh, and Nat1) and then normalized vs. control condition. Each dot represents an independent sample, n ≥ 3, with means ± SEM. The differentiation level at 72 h (bars control) is normalized to 100% to evaluate the reduction induced by 1C-MIM, where differences compared to control are significant at ∗∗∗p < 0.001.
(C and D) Effects on the supplementation of 1C-MIM over the progenitor markers of myofibers Pax7 and MyoD. Protein expression levels were evaluated by immunofluorescence in myofibers exposed to different concentrations of serum; differences compared to control are significant at ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.001.
(E) Representative images of in vitro control myofibers and those 3 days after the treatment with 1C-MIM. Note: a fresh feeding was done to improve the quality of the photograph; scale bar: 50 μm.
(F–J) Transcriptional characterization of the identity acquired by myofibers after 1C-MIM supplementation. (F) PCA map of control untreated myofibers, myofibers treated with a scramble combination of metabolites non-related directly to 1C (see main text for details), and 1C-MIM-treated myofibers. Transcriptomic profile obtained by bulk RNA-seq. (G) Hierarchical cluster analysis (HCA) performed using a Euclidean distance metric comparing gene expression profiles; red to blue color gradient indicates higher to lower expression. (H) Gene set enrichment analyses of the DEGs between 1C-MIM myofibers vs. the rest (control and scramble); to the left are enrichments for pathways, and to the right is enrichment for ontologies. (I) Volcano plot representations of differentially expressed genes (DEGs) at the comparison myofibers control to 1C-MIM. DEGs are represented by dots (adjusted p value [p-adj.] < 0.01). Blue dots indicate significantly upregulated and downregulated genes. Gray dots indicate not significant genes. (J) Enrichment analysis by gProfiler of DEGs included 471 downregulated and 351 upregulated considering p-adj. < 0.01.
See also Figures S2–S4 and Tables S2–S4.