Figure 6.
Modifications in acetylation and modulations on cell cycle drive the acceleration of regeneration by 1C-MIM in muscle in vivo
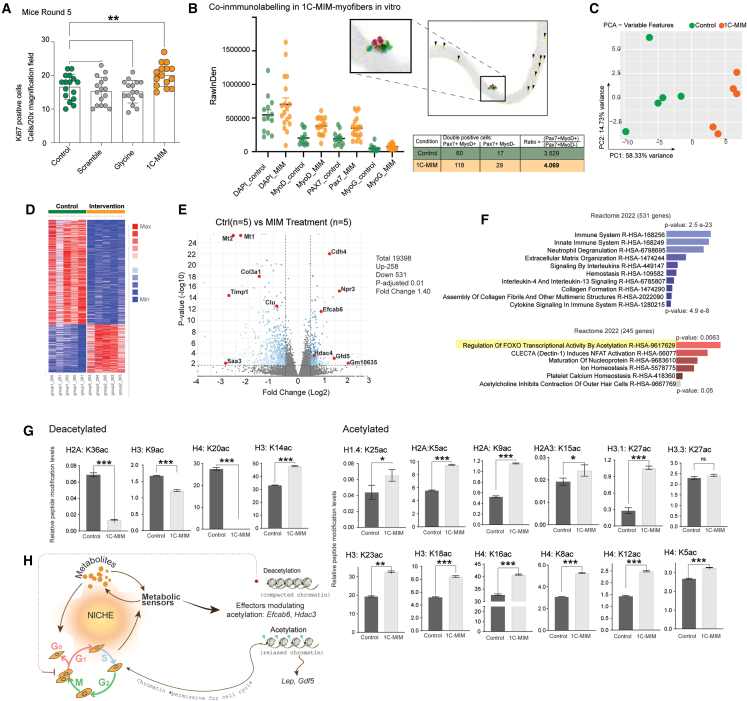
(A) Proliferation quantified by immunofluorescence and Ki67 detection.
(B) Primary myofibers were placed in vitro and treated with or without 1C-MIM for precise evaluation of double markers in nuclei. On the right, there is a summary table indicating the number of nuclear markers in single myofibers for control and 1C-MIM-treated double-positive and the ratio of Pax7+ vs. MyoD+ or MyoD– in myofibers exposed to the indicated conditions.
(C–F) Transcriptomic analysis of 1C-MIM quadriceps extracted from old mice after 3 months of supplementation. (C) PCA. (D) Heatmap with the total of DEGs. (E) Respective volcano plots showing DEGs as blue dots DEG, not significant genes as gray dots, and genes of interest highlighted as red dots (p-adj. <0.01). (F) Enrichment analysis of the DEGs with p-adj. < 0.01.
(G) Orthogonal readouts on the histone modifications detected by mass spectrometry on control astrocytes and 1C-MIM-supplemented astrocytes.
(H) Suggested mechanism of 1C-MIM for eliciting acceleration of the cell transition on regeneration. Several metabolic sensors respond to incoming metabolites, creating modulation feedback in the niche. Particularly, an increase in deacetylation will evoke relaxed chromatin, facilitating the expression of new genes associated with specific cell programs required for a cell in a given time (e.g., during regeneration). Thus, preloading the cells with specific metabolites may have prepared them for the rapid gene activation required for any identity transition.
Total intervention n = 87 mice. Pair comparisons at indicated groups are in Figure S6A. See also Figure S7.
Significant differences as ∗p<0.05, ∗∗p<0.005, and ∗∗∗p<0.001.