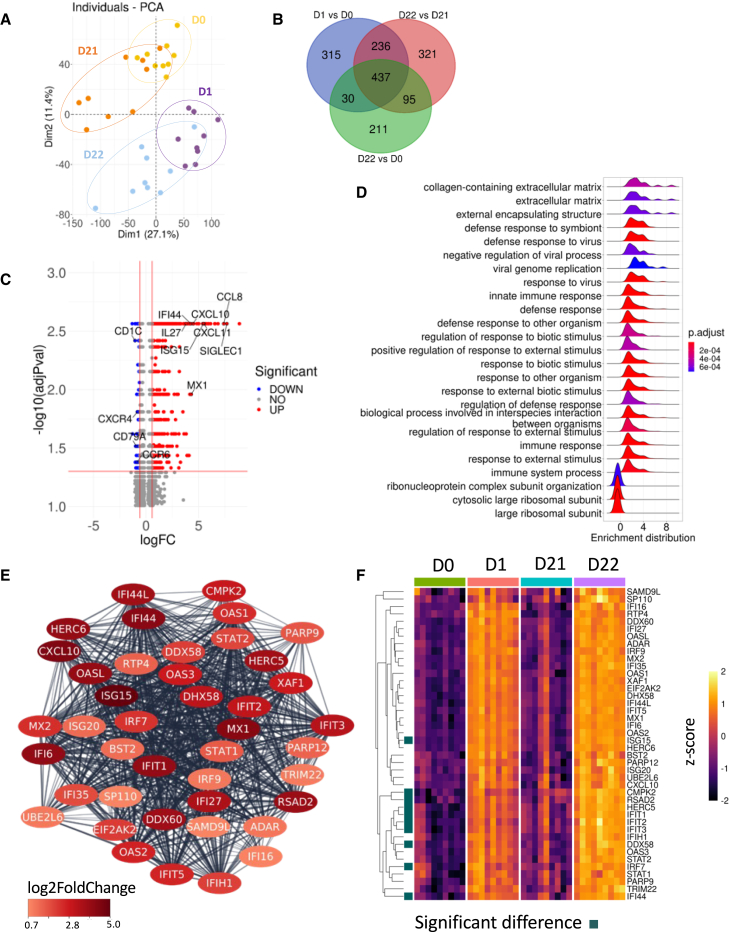
Figure 7.
Characterization of gene expression profiles induced after prime-boost vaccinations
(A) Principal-component analysis based on the full transcriptomic profile of vaccinated AGMs sampled at day 0 (before vaccination) (yellow); day 1 after prime vaccination (purple); 1 day before the boost, corresponding to day 21 (orange); and 1 day after the boost (day 22) (sky blue).
(B) Venn diagram of differential expression of genes (DEGs) comparing VAC day 1 to 0, 22 to 21, and 22 to 0 (adjusted p value ≤ 0.05, absolute log2 fold change [FC] ≥ 0.58).
(C) Volcano plot of DEGs observed between VAC day 22 and 0. The down- (190) and upregulated (583) genes are shown in blue and red, respectively. The top 35 most upregulated and down-regulated genes based on the log2FC are depicted.
(D) Gene set enrichment analysis was performed on the DEGs with an adjusted p value ≤ 0.05 between VAC day 22 and 0 using the ClusterProfiler v.4.4.4 R package. The density plot shows the top 25 pathways based on the adjusted p value and normalized enrichment score.
(E) The highest ranked network of DEGs (adjusted p value ≤ 0.05, absolute log2FC ≥ 0.58) between VAC days 22 and 0 was obtained from the protein-protein interaction (PPI) network using the MCODE plug-in of Cytoscape. The DEGs in the network are colored based on the log2FC values.
(F) Heatmap of gene expression belonging to the MCODE network at days 0 (green), 1 (salmon), 21 (blue), and 22 (purple). Gene expression levels in the heatmap are shown as Z scores. Significantly differentially expressed genes in post-boost versus post-prime are indicated by a square.