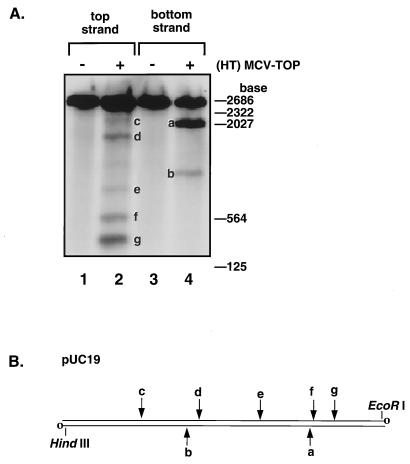
FIG. 3.
Site-specific cleavage by (HT)MCV-TOP. (A) Linear pUC19 DNA uniquely labeled on each 3′ end was incubated with purified MCV topoisomerase, treated with 0.5% SDS, and separated on 1.6% alkaline agarose gels by electrophoresis. Lanes: 1, no topoisomerase; 2, 400 ng of topoisomerase; 3, no topoisomerase; 4, 400 ng of topoisomerase. To generate the cleavage substrate, pUC19 DNA was linearized with XbaI and 3′ end labeled with Klenow enzyme and [α-32P]dCTP. Linear pUC19 labeled on one end only was then prepared by digestion with EcoRI (top strand) or HindIII (bottom strand) to remove the label at the undesired position. Reaction mixtures (20 μl) containing 20 mM Tris-HCl, pH 8.0, 200 mM K-glutamate, 1 mM dithiothreitol, 0.1% Triton X-100, 1 mM EDTA, and 50 ng of 3′ end 32P-labeled linear pUC19 DNA plus 400 ng topoisomerase were incubated at 37°C for 5 min. Cleaved DNA products were disrupted by the addition of SDS to 0.5%, heated at 95°C for 5 min, cooled on ice, adjusted to 50 mM NaOH and 1 mM EDTA, and then analyzed by electrophoresis on a 1.6% alkaline agarose gel (16). (B) Diagram of the cleavage substrate and the approximate positions of MCV topoisomerase cleavage sites in pUC19. The site of strand labeling in each set of reactions is indicated by open circles. The cleavage sites are indicated on the top or bottom strand by arrows.