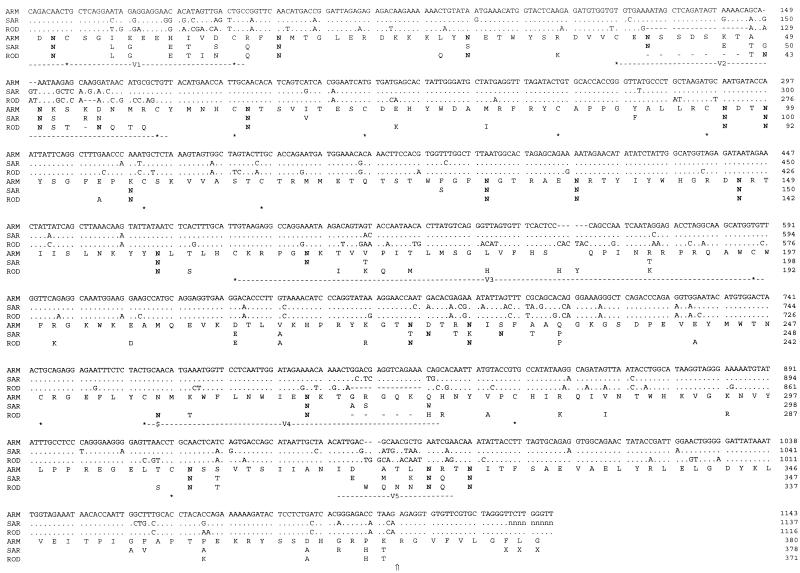
FIG. 1.
Alignment of HIV-2 strain ARM, SAR, and ROD nucleotide and translation product sequences spanning the gp125 region. For the nucleotide sequences, dashes indicate gaps introduced to improve the alignment, dots show agreement with the ARM sequence, and the letter n represents nucleotides not resolved in the SAR sequence. For amino acid alignments, residues which differ from the ARM sequence are shown, dashes mark deletions compared to the ARM sequence, and X indicates an amino acid that cannot be assigned in the SAR translation. The locations of asparagine residues (N) which form part of a potential N-linked glycosylation sequon are in boldface type, and $ marks the position of the deleted sequon in the ARM and SAR sequences. The positions of conserved cysteines are indicated (asterisks), as are the locations of the variable domains (V1 to V5) and the cleavage site between gp125 and gp41 (⇑).