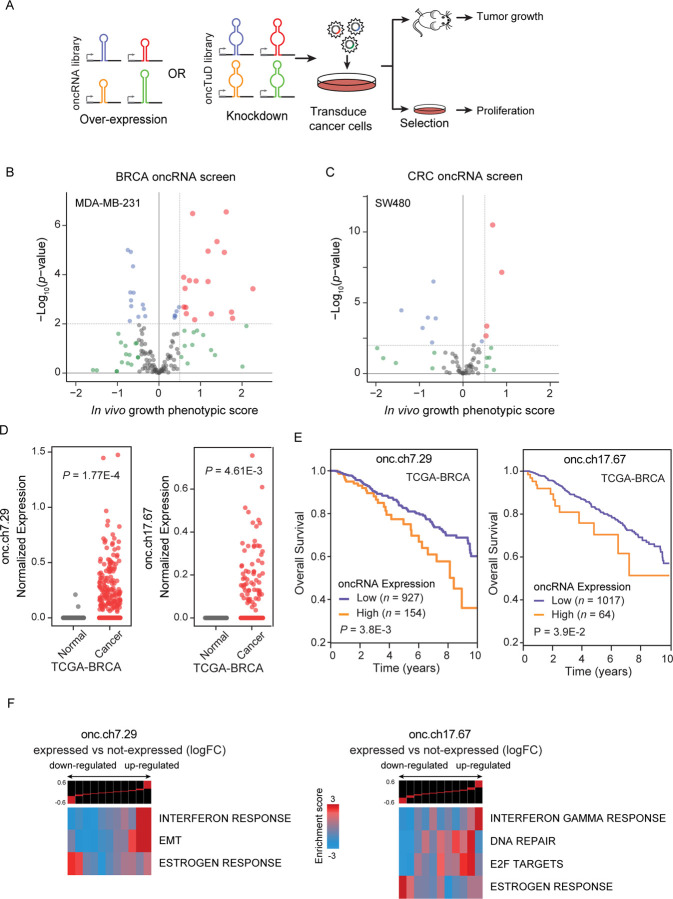
Figure 3. Systematic annotation of driver oncRNAs using a scalable in vivo genetic screening approach.
(A) Workflow schematic of oncRNA cancer and oncRNA TuD functional screens. (B-C) Volcano plots of oncRNA functional screen results for breast cancer (MDA-MB-231) and colorectal cancer (SW480), respectively. In vivo growth phenotypic score refers to enriched representation of cancer cells transduced with cognate oncRNA upon tumor growth in the xenograft model. (D) Expression levels of two example oncRNAs with significant tumor growth phenotype from the functional screen in TCGA-BRCA tumor and tumor-adjacent normal tissues. P values were calculated using a one-tailed Mann-Whitney test. (E) Survival of TCGA-BRCA patients stratified by expression level of cognate driver oncRNA. P values were calculated using a log-rank test. (F) Informative iPage pathways associated with TCGA-BRCA cancer samples expressing cognate oncRNAs compared to TCGA-BRCA cancer samples with no detectable respective oncRNAs. Top panel shows gene expression differences in discrete expression bins. Genes that are up-regulated in oncRNA expressing cancer samples are in the right bins, whereas bins to the left contain genes with lower expression. The heatmap shows the corresponding pathway in relation to the expression bins. Red entries indicate enrichment of pathway genes in a given expression bin whereas blue entries indicate depletion. Enrichment and depletion are measured using log-transformed hypergeometric P values.