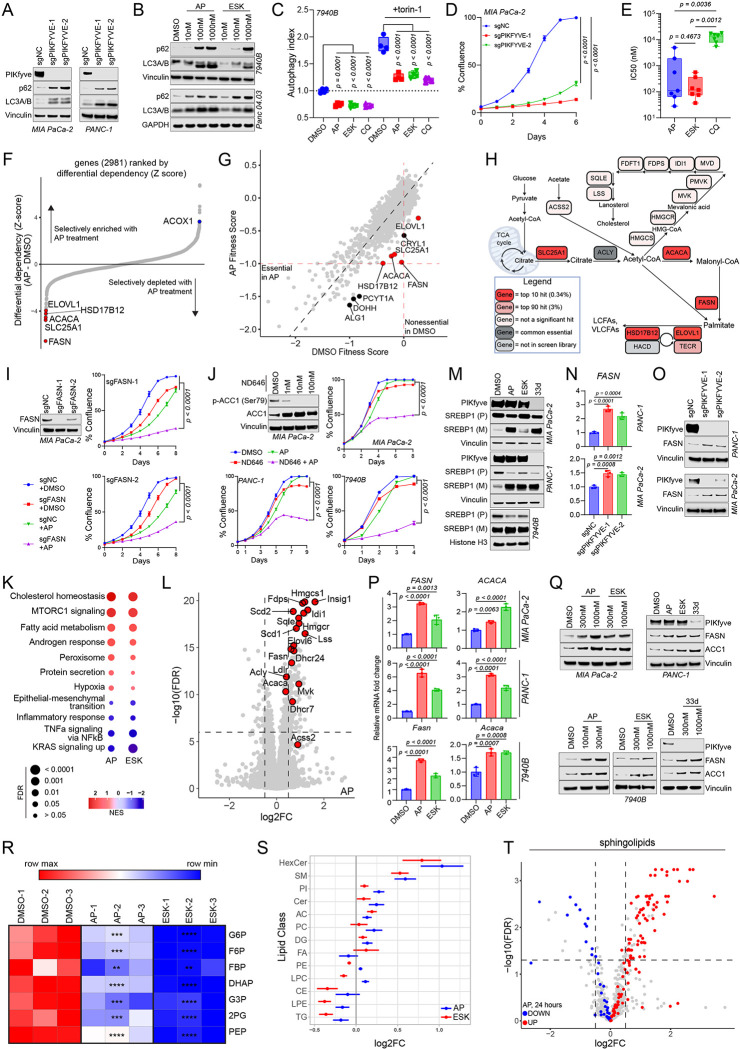
Figure 3: PIKfyve inhibition obligates PDAC cells to stimulate a lipogenic transcriptional and metabolic program.
A. Immunoblot analysis of MIA PaCa-2 (left) and PANC-1 (right) cells upon CRISPRi-mediated knockdown of PIKFYVE with two independent sgRNAs (sgPIKFYVE-1 and sgPIKFYVE-2) or control (sgNC) showing changes in PIKfyve, p62 (SQSTM1), and LC3A/B. Vinculin was used as a loading control for all blots. This experiment was performed twice independently with similar results.
B. Immunoblot analysis of known autophagy markers p62 (SQSTM1) and LC3A/B upon treatment with PIKfyve inhibitors apilimod (AP) or ESK981 (ESK) in KPC 7940B and Panc 04.03 cell lines. Vinculin or GAPDH were used as loading controls. The immunoblot using 7940B cells was performed twice independently with similar results.
C. Tandem fluorescent autophagic flux reporter assay in 7940B cells after 24-hour treatment with apilimod (100 nM), ESK981 (1000 nM), and chloroquine (CQ, 50 μM) with or without mTORC1/mTORC2 inhibitor torin-1 (100 nM). Data shown are 4 biological replicates for each condition. (One-way ANOVA with Dunnett’s using indicated conditions as baseline). This represents one of three independent experiments.
D. Confluence assay of MIA PaCa-2 cells upon CRISPRi-mediated knockdown of PIKFYVE (sgPIKFYVE) or control (sgNC). Data shown are mean +/− SEM (n=4 biological replicates) (Two-way ANOVA with Dunnet’s). This represents one of three independent experiments.
E. Box-and-whisker plot displaying IC50s of apilimod, ESK981, and chloroquine in 7 human and mouse PDAC cell lines (specified in Extended Data Fig. 4 B–E). Statistics were performed using a Repeated Measures one-way ANOVA with Reisser-Greenhouse correction and with Tukey’s multiple comparisons test with individual variances computed for each comparison.
F. Gene enrichment rank plot based differential sgRNA representation in apilimod-treated versus DMSO-treated endpoint populations of the CRISPR screen experiment. Lipid synthesis-related genes ranked at either extreme are highlighted.
G. Scatter plot of gene fitness scores in apilimod-treated versus DMSO-treated endpoint conditions in metabolic CRISPR screen. Top 10 hits are labeled, and 5 lipid synthesis-related genes are highlighted.
H. Metabolic map of fatty acid synthesis and elongation, and cholesterol homeostasis. Red indicates top 10 hit in the CRISPR screen; pink indicates top 90 (3%) hit; light pink indicates the gene was not a top 3% hit; dark grey indicates the gene is universally essential; light grey indicates the gene was not included in the CRISPR screen library.
I. Immunoblot of MIA PaCa-2 upon CRISPRi-mediated knockdown of FASN in cells and corresponding confluence assays assessing the sensitivity of FASN knockdown (sgFASN) or control (sgNC) cells to apilimod (100 nM). Vinculin was used as a loading control for the immunoblot. Confluence assay data shown are mean +/− SEM (n=4) from one of two independent experiments. Statistics were performed using an F statistics test based on a two-way ANOVA with Dunnett’s multiple comparisons test with the sgFASN + apilimod condition set as a baseline.
J. Immunoblot analysis assessing the phosphorylation status of ACC1 (p-ACC1) in MIA PaCa-2 cells upon ND646 (ACC inhibitor) treatment and corresponding confluence assays assessing the sensitivity of MIA PaCa-2, PANC-1, and 7940B cells to apilimod, ND646, or both. Vinculin was used as a loading control for the immunoblot. Confluence assay data shown are mean +/− SEM (n=4). Statistics were performed using a two-way ANOVA with Dunnett’s multiple comparisons test with the ND646+apilimod condition set as a baseline. Concentrations used for apilimod are: 100 nM for MIA PaCa-2 and 50 nM for PANC-1 and 7940B. Concentrations used for ND646 are: 100 nM for MIA PaCa-2 and 1000 nM for PANC-1 and 7940B. The confluence assays were performed three times independently with similar results. The DMSO and ND646 conditions for MIA PaCa-2 and PANC-1 are also utilized as controls in Extended Data Fig. 6F and G as these data were generated from the same experiment.
K. Pathway enrichment analysis of RNA-seq performed on 7940B treated with either apilimod (25 nM) or ESK981 (250 nM) for 8 hours. Dot sizes are inversely proportional to false discovery rate (FDR). The color scheme is reflective of the normalized enrichment score (NES).
L. Volcano plot using RNA-seq analysis on 7940B cells treated with apilimod (25 nM) for 8 hours highlighting SREBP-1 target genes. Vertical dashed lines indicate log2 fold change = +/− 0.5). Horizontal dashed line indicates FDR=10−6.
M. Immunoblot showing PIKfyve, premature SREBP1 (SREBP1 (P)), and mature SREBP1 (SREBP1 (M)) in MIA PaCa-2, PANC-1, and 7940B cells upon treatment with PIKfyve inhibitors or degrader PIK5–33d (33d) for 8 hours. Vinculin or histone H3 were used as loading controls. The drug doses used were as follows: MIA-PaCa-2 and PANC-1: apilimod=300 nM, ESK981=1000 nM, PIK5–33d=1000 nM; 7940B: apilimod=100 nM, ESK981=1000 nM, PIK5–33d=1000 nM. This data is representative of two independent experiments.
N. Quantitative-PCR (qPCR) of MIA PaCa-2 and PANC-1 showing changes in RNA levels of FASN upon CRISPRi-mediated knockdown of PIKFYVE using two independent sgRNAs targeting PIKFYVE compared to control. Data plotted are technical triplicates from one of three independent experiments. (One-way ANOVA with Dunnett’s).
O. Immunoblot analysis of MIA PaCa-2 and PANC-1 showing changes in protein levels of FASN upon CRISPRi-mediated knockdown of PIKFYVE using two independent sgRNAs targeting PIKFYVE relative to control. Vinculin was used as a loading control. These data are representative of two independent experiments.
P. qPCR of MIA PaCa-2, PANC-1, and 7940B showing changes in RNA levels of labeled genes upon treatment with PIKfyve inhibitors for 8 hours. The drug doses used were as follows: MIA PaCa-2 and PANC-1: apilimod = 300 nM, ESK981 = 1000 nM. 7940B: apilimod = 100 nM, ESK981 = 1000 nM. Data plotted are technical triplicates (One-way ANOVA with Dunnett’s). These experiments were performed three independent times each with similar results.
Q. Immunoblot analysis of MIA PaCa-2, PANC-1, and 7940B showing changes in protein levels of labeled genes upon treatment with PIKfyve inhibitors for 24 hours. Vinculin was used as a loading control. The drug doses used were indicated on figure or as follows for PANC-1: apilimod = 300 nM, ESK981 = 1000 nM, PIK5–33d = 1000 nM. These data are representative of two independent experiments each.
R. Heatmap of glycolytic metabolite abundance in 7940B cells treated with DMSO, apilimod (100 nM), or ESK981 (1000 nM) for 8 hours. G6P = glucose 6-phosphate; F6P = fructose 6-phosphate; FBP = fructose 1,6-bisphosphate; DHAP = dihydroxyacetone phosphate; G3P = glyceraldehyde 3-phosphate; 2PG = 2 phosphoglycerate; PEP = phosphoenolpyruvate. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001. (One-way ANOVA with Dunnett’s)
S. Forest plot indicating changes in lipid class abundance in 7940B cells upon treatment with DMSO, apilimod (100 nM) or ESK981 (1000 nM) for 24 hours. HexCer = hexosylceramide; SM = sphingomyelin; PI= phosphatidylinositol; Cer = ceramide; AC= acylcarnitine; PC= phosphatidylcholine; DG = diacylglyceride; FA = fatty acid; PE = phosphatidylethanolamine; LPC = lysophosphatidylcholine; CE = cholesteryl ester; LPE = lipophosphatidylethanolamine; TG = triacylglyceride. Effect sizes are in log2 scale of lipid abundance estimated from separate linear model for each treatment (apilimod or ESK981) compared to DMSO, adjusting for lipid classes with random intercept.
T. Volcano plot of lipidomics analysis on 7940B cells treated with apilimod for 24 hours, plotting log2 fold change compared to DMSO highlighted changes in sphingolipid classes (HexCer, SM, Cer). Red highlights indicate upregulated sphingolipids; blue highlights indicate downregulated sphingolipids. Vertical dashed lines indicate log2 fold change = +/− 0.5). Horizontal dashed line indicates p = 0.05. (unpaired two-tailed t-test)