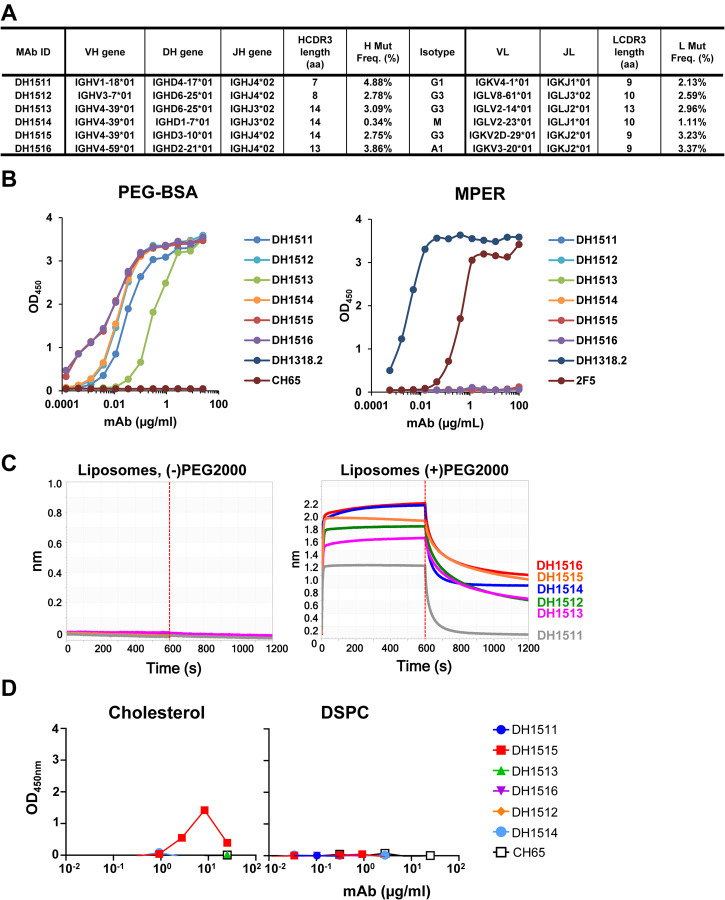
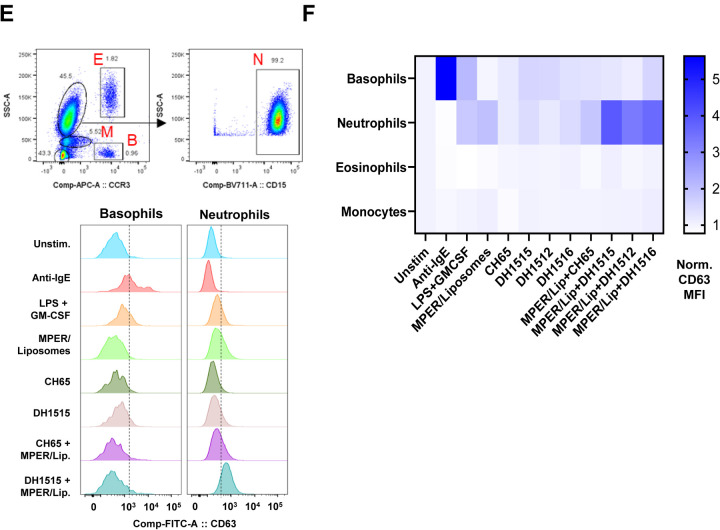
Figure 4. Induction of Anti-PEG IgG Responses in Recipients of MPER-peptide Liposome Vaccine.
(A) Immunogenetics of the six PEG-specific memory B cells recovered from recipient 133–33. (B) Six PEG-specific memory B cells were incidentally isolated from recipient 133–33 during B cell sorts with MPR03 antigen baits. When expressed as recombinant IgG1s, these antibodies bound to PEG (left) but not MPER (right), indicating that these mAbs were not cross- or poly-reactive. (C) PEG-specific mAbs were tested for binding liposomes without (left) and with (right) PEG in BLI. Binding responses were measured in nm. (D) PEG-specific mAbs were tested for binding cholesterol and lipids (DSPC) via ELISA. Binding was measured as OD450nm. (E) A modified Basophil Activation Test was used to test the capacity of anti-PEG IgG1 mAbs to sensitize basophils and other myeloid cells to MPER peptide liposomes. Flow cytometric gating strategy to identify basophils (B), eosinophils (E), monocytes (M), and neutrophils (N) among peripheral blood leukocytes. Histograms show CD63 labeling of basophils and neutrophils after incubation with stimulatory agents anti-IgE or LPS+GM-CSF, or with combinations of different mAbs, including CH65 mAb (a control anti-flu HA IgG1) and DH1515 mAb (anti-PEG IgG1), with or without co-culture with MPER peptide liposome vaccine material. (F) Heat map quantification of CD63 labeling of basophils, neutrophils, eosinophils, and monocytes after incubation with stimulatory agents (anti-IgE, LPS+GM-CSF), control IgG1 mAb (CH65) with or without co-incubation with MPER peptide liposome vaccine, or anti-PEG IgG1 mAbs (DH1512, DH1515 and DH1516) with our without MPER/Liposome vaccine material. Data for each condition represent CD63 MFI normalized to that in the unstimulated condition. Data represent one of three experimental replicates.