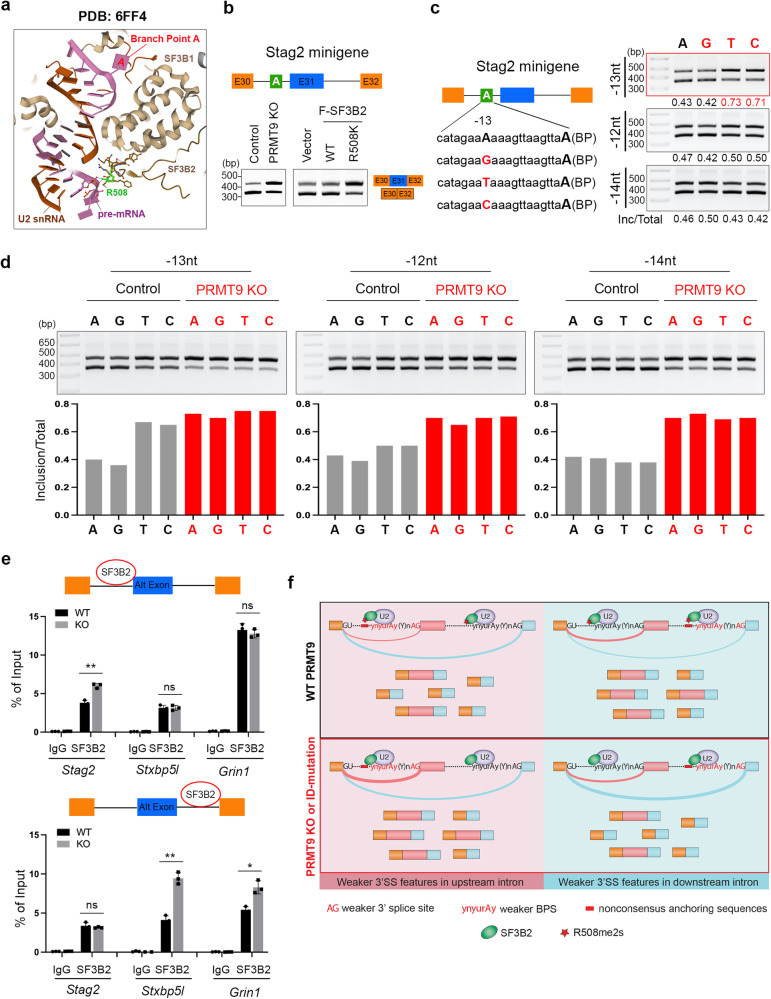
Fig. 7. PRMT9-mediated SF3B2 R508me2s regulates splicing through the pre-mRNA anchoring site.
a Cryo-EM structure of the human spliceosome complex (PDB: 6FF4). Branch point on the pre-mRNA is shown in red. SF3B2 R508 is highlighted in green. b A schematic illustration of the Stag2 splicing minigene. The alternative exon 31 is labeled in blue. The anchoring site is highlighted in green (upper panel). RT-PCR was performed to detect the alternative splicing products in control and PRMT9 KO HEK293 cells, as well as in cells expressing WT and R508K mutant SF3B2 (lower panel). n = 3. c Single nucleotide variations at the anchoring site differentially regulate Stag2 minigene splicing. WT and mutant Stag2 splicing minigenes that contain single nucleotide mutations at −12, −13, and −14 nt upstream of the BP were compared by RT-PCR. The relative ratios of inclusion vs total (Inc/Total) were quantified using ImageJ software (n = 3). d PRMT9 KO promotes exon inclusion and renders splicing to be insensitive to −13nt nucleotide variations. The splicing patterns of Stag2 minigenes containing WT or single nucleotide mutations at −12, −13, and −14 nt upstream of the BP were detected in control and PRMT9 KO HEK 293 T cells by RT-PCR (n = 3). e PRMT9 negatively regulates SF3B2 interaction with nonconsensus anchoring sites. CLIP-qPCR was performed with the hippocampus tissues from WT and Prmt9 KO mice using the SF3B2 antibody. The amount of SF3B2-bound RNA was quantified by qPCR. Data are presented as mean ± SD (n = 3). *, p < 0.05; **, p < 0.01, ns, not significant. f Proposed working model: PRMT9-mediated SF3B2 R508me2s regulates RNA splicing through modulating SF3B2–anchoring site interaction and 3’ splice site selection. Source data are provided as a Source Data file.