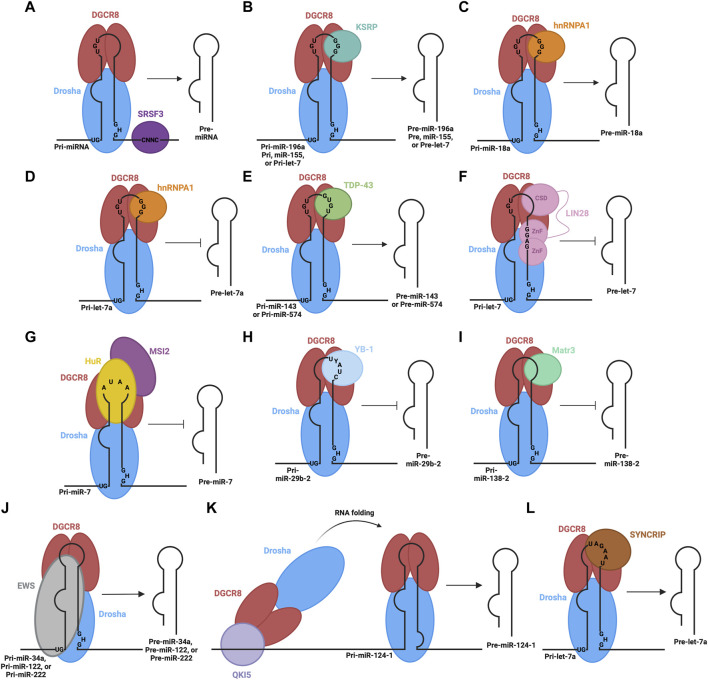
FIGURE 2.
RBPs involved in the processing of pri-miRNA in the nucleus. The microprocessor complex, consisting of Drosha and DGCR8 proteins, associates with pri-miRNA through their RNA binding domains and recognition of sequence motifs. Different RBPs also associate with different sequence elements of pri-miRNA: (A) SRSF3 promotes microprocessor-mediated pri-miRNA cleavage, (B) KSRP promotes microprocessor-mediated cleavage of pri-miR-196a, pri-miR-155, and pri-let-7, (C) hnRNPA1 promotes microprocessor-mediated cleavage of pri-miR-18a, (D) hnRNPA1 hinders microprocessor-mediated cleavage of pri-let-7a, (E) TDP-43 promotes microprocessor-mediated cleavage of pri-miR-143 and pri-miR-574, (F) LIN28B inhibits microprocessor-mediated cleavage of pri-let-7, (G) HuR and MSI2 inhibit microprocessor-mediated cleavage of pri-miR-7, (H) YB-1 inhibits microprocessor-mediated cleavage of pri-miR-29b-2, (I) Matr3 inhibits microprocessor-mediated cleavage of pri-miR-138-2, (J) EWS promotes microprocessor-mediated cleavage of pri-miR-34a, pri-miR-122, and pri-miR-222, (K) QKI5 promotes microprocessor-mediated cleavage of pri-miR-124-1 through recruitment to a distal QRE sequence and subsequent RNA folding to bring the microprocessor to the appropriate location, (L) SYNCRIP promotes microprocessor-mediated cleavage of pri-let-7a.