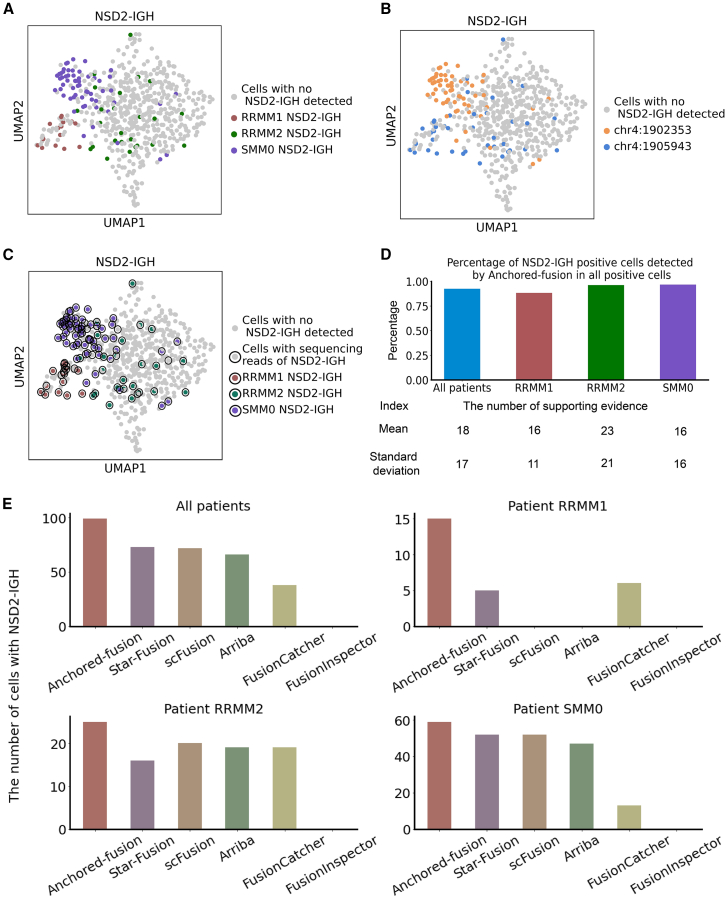
Figure 6.
Fusion detection using different tools for scRNA-seq dataset from a clinical cohort of patients with multiple myeloma
(A) UMAP visualization of MM scRNA-seq data. The single cells identified by Anchored-fusion as having the NSD2-IGH fusion gene in each patient are highlighted.
(B) UMAP visualization of MM scRNA-seq data. The single cells identified by Anchored-fusion with different breakpoints of NSD2 are highlighted.
(C) UMAP visualization of MM scRNA-seq data. All cells containing NSD2-IGH fusion gene are circled. The highlighted cells indicate those detected by Anchored-fusion tool from three NSD2-IGH-positive patients. We identified the cells containing the NSD2-IGH fusion gene with its reference sequence.
(D) The percentage of cells positive for NSD2-IGH detected by Anchored-fusion among all NSD2-IGH-positive cells. In the lower part of the image are the indices of the number of the support evidences per 1 million reads in NSD2-IGH-positive cells detected by Anchored-fusion.
(E) Bar charts showing the total number of cells containing the NSD2-IGH fusion gene detected by Anchored-fusion and five other methods in the MM single-cell dataset, along with the number of cells detected in each patient.