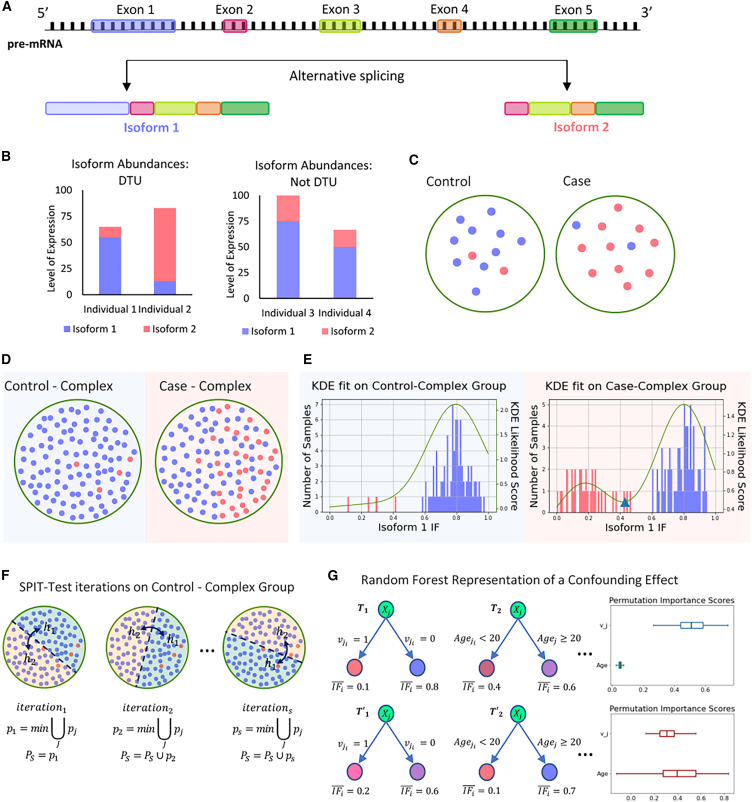
Figure 1.
DTU detection demonstration
(A) Gene locus going though alternative splicing to produce isoform 1 and isoform 2.
(B) Left: isoform abundances in a sample case of DTU between individuals 1 and 2. Right: isoform abundances in a sample case without DTU but with changes in overall expression between individuals 3 and 4.
(C) Conventional DTU analysis assumption with no structured heterogeneity in either group.
(D) Heterogeneity structure in complex disease samples, where a subset of cases shares the same genetic abnormality (case-complex).
(E) Corresponding isoform fraction (IF) distributions and KDE fits for the samples represented in groups control-complex and case-complex.
(F) Three SPIT-Test iterations demonstrated with random splits of the control-complex group.
(G) Random forest regression representation when there is not a significant confounding effect in the DTU transcript (top) vs. when there is a clear confounding effect by the covariate “age” (bottom). Corresponding permutation importance scores for age and are shown on the right.
Samples (dots) are color coded based on their dominant isoforms for the locus in (C)–(F). Blue, isoform 1; red, isoform 2.