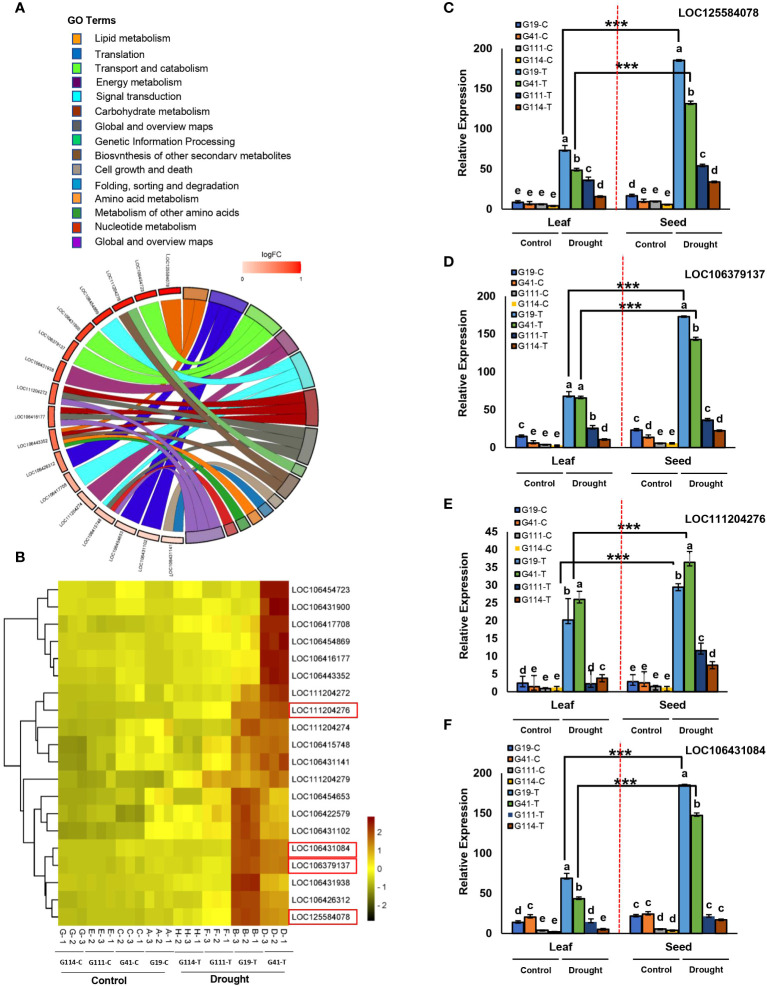
Figure 7.
Analysis of transcriptomes between high- and low-yielding varieties selected in drought stress treatment in the field study. Transcriptome study for days to ripening (DTR). (A) Detailed analysis of 15 enriched gene ontology groups selected using Circos plots for candidate genes in chromosomes A01, C03, and C06 related to DTR. Symbols of DEG from each of the analyzed comparisons are displayed on the left side of the graph with their logFC values, mapped by color scale (dark red = higher expression; light red = lower expression). Colored connecting lines determine gene involvement in the GO terms. (B) Heat map of the expression of 20 genes associated with DTR in chromosomes A01, C03, and C06 among low-yield varieties (G111 and G114) and high-yield varieties (G19 and G41) under drought stress. The red box indicates the key genes in the associated region of C03 related to DTR. Heatmap color represents the expression level of each gene (rows) under drought treatments (fold change >1, P-value <0.05). Red bars: upregulation; green bars: downregulation; (C): control and (T): drought treatment. (C-F) Tissue-specific expression of seven candidate genes in the peak regions of C03 from four varieties of rapeseed (low-yield varieties: G111 and G114; high-yield varieties: G19 and G41) under drought stress; (C): control and (T): drought treatment. Tissue-specific expression of the gene LOC125584078 (C), LOC106379137 (D), LOC111204276 (E), and LOC106431084 (F) between low-yield varieties and high-yield varieties under drought stress. Data are means ± SD, P-value < 0.05, as determined by multiple comparison testing by one-way ANOVA. Different letters indicate distinct groups. Red dotted line indicates division expression of genes between seed and leaf. Asterisks indicate significant difference between seed and leaf (Student’s t-test, *** P-value < 0.001).