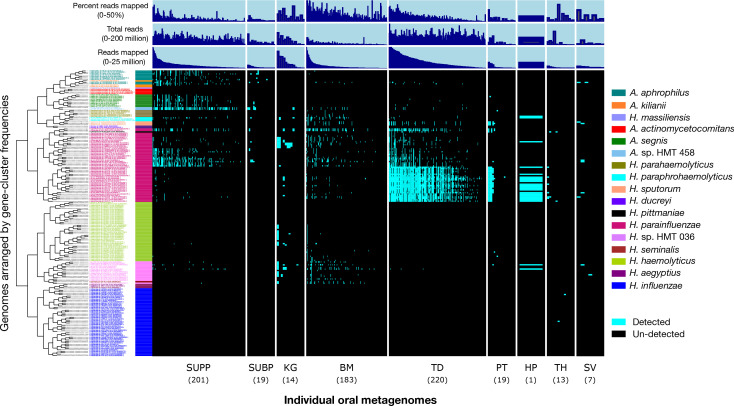
Fig 3.
Detection plot of Haemophilus and Aggregatibacter species and strains in 686 Human Microbiome Project (HMP) metagenomic samples from nine major oral sites. Each row displays the detection of a genome across all samples. A genome is detected (cyan) if at least 50% of its nucleotides have at least 1X coverage. If the genome is not detected in a sample, it is represented by a black bar. Samples are ordered by oral site and then by the decreasing number of reads mapped to the set of genomes. From left to right, oral sites are supragingival plaque (SUPP; n = 210), subgingival plaque (SUBP; n = 19), keratinized gingiva (KG; n = 14), (TD; n = 220 samples), palatine tonsil (PT; n = 19), throat (TH; n = 13), saliva (SV; n = 7), hard palate (HP; n = 1), and buccal mucosa (BM; n = 183). Additional data are shown for “total reads” in each sample, the number of “reads mapped” per sample, and “percent reads mapped” per sample.