ABSTRACT
Carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter spp. represent major threats and have few approved therapeutic options. Non-fermenting Gram-negative isolates were collected from hospitalized inpatients from 49 sites in 6 European countries between 01 January 2020 and 31 December 2020 and underwent susceptibility testing against cefiderocol and β-lactam/β-lactamase inhibitor combinations. Meropenem-resistant (MIC >8 mg/L), cefiderocol-susceptible isolates were analyzed by PCR, and cefiderocol-resistant isolates were analyzed by whole-genome sequencing to identify resistance mechanisms. Overall, 1,451 (950 P. aeruginosa; 501 Acinetobacter spp.) isolates were collected, commonly from the respiratory tract (42.0% and 39.3%, respectively). Cefiderocol susceptibility was higher than β-lactam/β-lactamase inhibitor combinations against P. aeruginosa (98.9% vs 83.3%–91.4%), and P. aeruginosa resistant to meropenem (n = 139; 97.8% vs 12.2%–59.7%), β-lactam/β-lactamase inhibitor combinations (93.6%–98.1% vs 10.7%–71.8%), and both meropenem and ceftazidime-avibactam (96.7% vs 5.0%–45.0%) or ceftolozane-tazobactam (98.4% vs 8.1%–54.8%), respectively. Cefiderocol and sulbactam-durlobactam susceptibilities were high against Acinetobacter spp. (92.4% and 97.0%) and meropenem-resistant Acinetobacter spp. (n = 227; 85.0% and 93.8%) but lower against sulbactam-durlobactam- (n = 15; 13.3%) and cefiderocol- (n = 38; 65.8%) resistant isolates, respectively. Among meropenem-resistant P. aeruginosa and Acinetobacter spp., the most common β-lactamase genes were metallo-β-lactamases [30/139; blaVIM-2 (15/139)] and oxacillinases [215/227; blaOXA-23 (194/227)], respectively. Acquired β-lactamase genes were identified in 1/10 and 32/38 of cefiderocol-resistant P. aeruginosa and Acinetobacter spp., and pirA-like or piuA mutations in 10/10 and 37/38, respectively. Conclusion: cefiderocol susceptibility was high against P. aeruginosa and Acinetobacter spp., including meropenem-resistant isolates and those resistant to recent β-lactam/β-lactamase inhibitor combinations common in first-line treatment of European non-fermenters.
IMPORTANCE
This was the first study in which the in vitro activity of cefiderocol and non-licensed β-lactam/β-lactamase inhibitor combinations were directly compared against Pseudomonas aeruginosa and Acinetobacter spp., including meropenem- and β-lactam/β-lactamase inhibitor combination-resistant isolates. A notably large number of European isolates were collected. Meropenem resistance was defined according to the MIC breakpoint for high-dose meropenem, ensuring that data reflect antibiotic activity against isolates that would remain meropenem resistant in the clinic. Cefiderocol susceptibility was high against non-fermenters, and there was no apparent cross resistance between cefiderocol and β-lactam/β-lactamase inhibitor combinations, with the exception of sulbactam-durlobactam. These results provide insights into therapeutic options for infections due to resistant P. aeruginosa and Acinetobacter spp. and indicate how early susceptibility testing of cefiderocol in parallel with β-lactam/β-lactamase inhibitor combinations will allow clinicians to choose the effective treatment(s) from all available options. This is particularly important as current treatment options against non-fermenters are limited.
KEYWORDS: cefiderocol, Pseudomonas aeruginosa, Acinetobacter spp., β-lactam/β-lactamase inhibitor combinations, meropenem, ceftazidime-avibactam, ceftolozane-tazobactam, meropenem-vaborbactam, imipenem-relebactam, aztreonam-avibactam, cefepime-taniborbactam, sulbactam-durlobactam, resistance, meropenem-resistant, β-lactamases, Europe, in vitro
INTRODUCTION
Antimicrobial resistance is widespread throughout Europe (1). In particular, the high rates of carbapenem resistance observed in Pseudomonas aeruginosa and Acinetobacter spp. (19% and 48%, respectively, in 2021) represent a major threat, considering few approved therapeutic options are available (1, 2). The World Health Organization has, therefore, recognized carbapenem-resistant (CR) P. aeruginosa and CR Acinetobacter baumannii (CRAB) as critical priority pathogens (2).
Major mechanisms of carbapenem resistance in P. aeruginosa include loss of outer membrane porin D function, overexpression of efflux pumps, and carbapenemases [including Class B metallo-β-lactamases (MBLs) such as the Verona integron-borne MBL (VIM) and emerging class A carbapenemases such as Guiana extended-spectrum β-lactamase (GES) and Klebsiella pneumoniae carbapenemase (KPC)] (3–9). Carbapenemases most commonly found in CR Acinetobacter spp. include OXA-23 and OXA-24/40 oxacillinases; with the exception of New Delhi MBL (NDM)-1, MBLs are less common, although their potency makes them problematic for treatment (5, 10–12). Efflux pumps and reduced membrane permeability due to CarO porin loss are also associated with carbapenem resistance in Acinetobacter spp. (13, 14).
Cefiderocol is a unique catechol-siderophore cephalosporin approved in Europe for the treatment of infections due to aerobic Gram-negative organisms in adults with limited treatment options (15). The mechanism of action of cefiderocol is the disruption of peptidoglycan cell-wall synthesis via inhibition of penicillin-binding proteins (PBPs) (16, 17). The structure of cefiderocol and “Trojan Horse” mechanism of bacterial cell entry provide enhanced stability to a wide range of β-lactamases, and allow for the activity of cefiderocol to be broadly unaffected by efflux pump overexpression and porin channel modifications observed in CR non-fermenters (17–21).
Various β-lactam/β-lactamase inhibitor (BLBLI) combinations have been approved (ceftazidime-avibactam, ceftolozane-tazobactam, imipenem-relebactam, and meropenem-vaborbactam) or are in development (aztreonam-avibactam, cefepime-taniborbactam, and sulbactam-durlobactam) for clinical use in Europe to treat infections caused by CR P. aeruginosa and/or Acinetobacter spp. (22–27). However, several of these combinations are affected or rendered inactive by one or more of the known mechanisms of resistance in CR non-fermenters; these BLBLI combinations remain unable to inhibit certain β-lactamases, MBLs in particular, and are still affected by porin channel modifications (28–32).
There is notable variation in the European Society of Clinical Microbiology and Infectious Diseases and Infectious Diseases Society of America guidance for treatment of infections due to CR P. aeruginosa and Acinetobacter spp., although both generally recognize that in vitro activity of antimicrobials is an important consideration for treatment decision-making (33, 34). The longitudinal surveillance studies SENTRY and SIDERO generated data on the susceptibilities of cefiderocol and approved BLBLI combinations against non-fermenters, including CR and BLBLI combination-resistant isolates (35, 36). Although, despite the current and anticipated use of cefiderocol and BLBLI combinations against CR Gram-negative infections, there remains limited data comparing these antimicrobials and developmental BLBLI combinations against CR non-fermenter isolates, including those also resistant to a comparator antimicrobial.
The aim of this study was to evaluate the in vitro activity of cefiderocol, meropenem, BLBLI combinations (approved and in development), and colistin against clinical Gram-negative isolates collected between 01 January 2020 and 31 December 2020, across six countries in Europe. Here, we report the results for P. aeruginosa and Acinetobacter spp. isolates. Results for Enterobacterales isolates collected in this study are reported elsewhere.
RESULTS
Epidemiology
In total, 1,451 isolates were collected from European hospitals, of which 950 (65.5%) were P. aeruginosa and 501 (34.5%) were Acinetobacter spp. [including 458 (91.4%) A. baumannii complex] (see Table S1 for isolates by country). P. aeruginosa and Acinetobacter spp. isolates were collected from a range of infection sources, the most common for both species being respiratory tract [42.0% (399/950); 39.3% (197/501)], bloodstream [25.4% (241/950); 26.3% (132/501)], and skin [21.7% (206/950); 20.0% (100/501)], respectively (see Fig. S1 for all infection sources).
Susceptibility profiles of isolates
Susceptibility to cefiderocol was 98.9% against P. aeruginosa isolates and 92.4% against Acinetobacter spp. isolates [Table 1; see Table S2 for susceptibility rates using Clinical and Laboratory Standards Institute (CLSI) breakpoints or United States Food and Drug Administration (FDA) breakpoints where CLSI breakpoints were not available]. By individual country, susceptibility to cefiderocol was similar against P. aeruginosa, ranging from 98.7% in France to 99.3% in the United Kingdom, but showed more variability against Acinetobacter spp. (88.7% in Italy to 98.0% in Spain) (Table S3A through E). Against P. aeruginosa isolates, rates of susceptibility to BLBLI combinations were lower than cefiderocol, ranging from 83.3% for imipenem-relebactam to 91.4% for cefepime-taniborbactam (Table 1). Against Acinetobacter spp. isolates, rates of susceptibility were 92.4% to cefiderocol and 97.0% to sulbactam-durlobactam (FDA breakpoint) (Table 1) (37, 38). Using epidemiological cut-off (ECOFF) values, susceptibility to colistin was 99.7% and 98.4% for P. aeruginosa and Acinetobacter spp., respectively (Table 1).
TABLE 1.
In vitro activity of cefiderocol, BLBLI combinations, and other relevant antibiotics against P. aeruginosa and Acinetobacter spp. isolatesa,b
| FDC | MEM | CZA | C-T | MVB | I-R | ATM-AVI | FEP-TAN | SUL-DUR | (CST) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates (n) | MIC90 (mg/L) | S (%) | MIC90 (mg/L) | S/I (%) | MIC90 (mg/L) | S (%) | MIC90 (mg/L) | S (%) | MIC90 (mg/L) | S/I (%) | MIC90 (mg/L) | S (%) | MIC90 (mg/L) | S/I (%) | MIC90 (mg/L) | S/I (%) | MIC90 (mg/L) | S (%) | MIC90 (mg/L) | S (%) |
|
P. aeruginosa (950) |
1 | 98.9 | 16 | 85.4 | 8 | 90.1 | 8 | 89.1 | 16 | 87.2 | 4 | 83.3 | 32 | 86.2 | 8 | 91.4 | N/A | N/A | (1) | (99.7) |
| Acinetobacter spp. (501)c | 2 | 92.4 | >16 | 54.7 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 4 | 97.0 | (0.5) | (98.4) |
ATM-AVI, aztreonam-avibactam; BLBLI, β-lactam/β-lactamase inhibitor; CST, colistin; C-T, ceftolozane-tazobactam; CZA, ceftazidime-avibactam; ECOFF, epidemiological cut-off; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FDA, Food and Drug Administration; FDC, cefiderocol; FEP-TAN, cefepime-taniborbactam; I, susceptible, increased exposure; I-R, imipenem-relebactam; MEM, meropenem; MVB, meropenem-vaborbactam; N/A, not applicable; PD, pharmacodynamic; PK, pharmacokinetic; S, susceptibility; SUL-DUR, sulbactam-durlobactam.
Antibiotics were tested against P. aeruginosa and/or Acinetobacter spp. based on expected use in a real-world setting. Susceptibility was assessed according to EUCAST breakpoints (including non-species-specific PK/PD breakpoints, high-dosage breakpoints, and breakpoints for the agent without inhibitor, where applicable), except for sulbactam-durlobactam and colistin where FDA breakpoints and ECOFF values were used, respectively. Data on susceptibility to colistin are shown in parentheses as colistin is not recommended for monotherapy and is not associated with a clinical monotherapy breakpoint (as per EUCAST v.14.0 guidance).
Includes 458 A. baumannii complex isolates.
Susceptibility profiles of isolates with antibiotic-resistant phenotypes
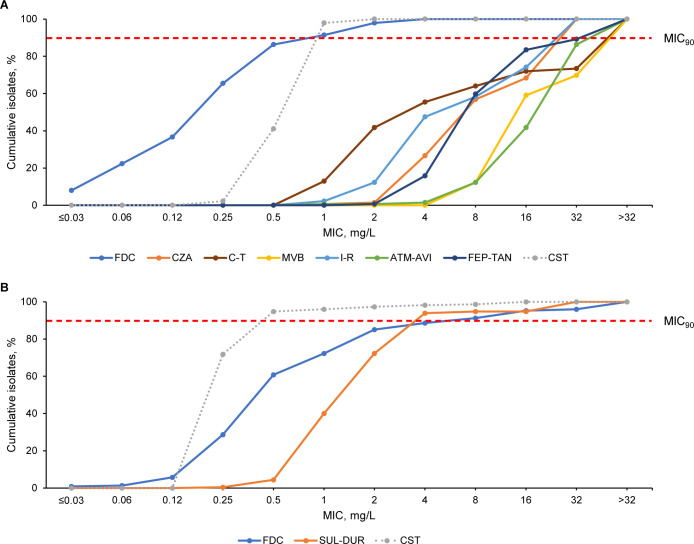
In total, 14.6% (139/950) P. aeruginosa isolates were classified as meropenem resistant (meropenem MIC >8 mg/L) (Table 2; see Fig. 1A for MIC distributions). Susceptibility of meropenem-resistant P. aeruginosa to cefiderocol was 97.8%, which was higher than susceptibility rates of <60% to all tested BLBLI combinations (Table 2). Meropenem-vaborbactam and imipenem-relebactam had the lowest susceptibility (12.2%), followed by aztreonam-avibactam (41.7%) and then cefepime-taniborbactam, ceftazidime-avibactam, and ceftolozane-tazobactam (55.4%–59.7%) (Table 2). This trend was detected across all countries (Table S4A through D).
TABLE 2.
In vitro activity of cefiderocol, BLBLI combinations, and other relevant antibiotics against P. aeruginosa and Acinetobacter spp. isolates with resistant phenotypesa,b
| Isolates | n | Susceptibilityc (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FDC | MEM | CZA | C-T | MVB | I-R | ATM-AVI | FEP-TAN | SUL-DUR | (CST) | ||
| P. aeruginosa | 950 | 98.9 | 85.4 | 90.1 | 89.1 | 87.2 | 83.3 | 86.2 | 91.4 | N/A | (99.7) |
| MEM-R | 139 | 97.8 | 56.8 | 55.4 | 12.2 | 12.2 | 41.7 | 59.7 | N/A | (100) | |
| CZA-R | 94 | 93.6 | 36.2 | 36.2 | 39.4 | 37.2 | 54.3 | 55.3 | N/A | (100) | |
| C-T-R | 104 | 94.2 | 40.4 | 42.3 | 45.2 | 36.5 | 64.4 | 59.6 | N/A | (100) | |
| MVB-R | 122 | 97.5 | 0 | 53.3 | 53.3 | 10.7 | 38.5 | 59.0 | N/A | (100) | |
| I-R-R | 159 | 98.1 | 23.3 | 62.9 | 58.5 | 31.4 | 54.1 | 64.8 | N/A | (100) | |
| ATM-AVI-R | 131 | 95.4 | 38.2 | 67.2 | 71.8 | 42.7 | 44.3 | 61.8 | N/A | (100) | |
| FEP-TAN-R | 82 | 95.1 | 31.7 | 48.8 | 48.8 | 39.0 | 31.7 | 39.0 | N/A | (100) | |
| MEM-R and CZA-R | 60 | 96.7 | 28.3 | 5.0 | 8.3 | 45.0 | 43.3 | N/A | (100) | ||
| MEM-R and C-T-R | 62 | 98.4 | 30.6 | 8.1 | 9.7 | 54.8 | 50.0 | N/A | (100) | ||
| Acinetobacter spp. | 501d | 92.4 | 54.7 | N/A | N/A | N/A | N/A | N/A | N/A | 97.0 | (98.4) |
| FDC-R | 38 | 10.5 | N/A | N/A | N/A | N/A | N/A | N/A | 65.8 | (92.1) | |
| MEM-R | 227 | 85.0 | N/A | N/A | N/A | N/A | N/A | N/A | 93.8 | (97.4) | |
| SUL-DUR-Re | 15 | 13.3 | 6.7 | N/A | N/A | N/A | N/A | N/A | N/A | (100) | |
ATM-AVI, aztreonam-avibactam; BLBLI, β-lactam/β-lactamase inhibitor; CST, colistin; C-T, ceftolozane-tazobactam; CZA, ceftazidime-avibactam; ECOFF, epidemiological cut-off; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FDA, Food and Drug Administration; FDC, cefiderocol; FEP-TAN, cefepime-taniborbactam; I-R, imipenem-relebactam; MEM, meropenem; MVB, meropenem-vaborbactam; N/A, not applicable; PD, T, ceftolozane-tazobactam; CZA, ceftazidime-avibactam; ECOFF, epidemiological cut-off; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FDA, Food and Drug Administration; FDC, cefiderocol; FEP-TAN, cefepime-taniborbactam; I-R, imipenem-relebactam; MEM, meropenem; MVB, meropenem-vaborbactam; N/A, not applicable; PD, pharmacodynamic; PK, pharmacokinetic; R, resistant; SUL-DUR, sulbactam-durlobactam.
Antimicrobials were tested against P. aeruginosa and/or Acinetobacter spp. based on expected use in a real-world setting. Results are not reported for isolates tested against antibiotics to which they had an expected resistance phenotype. Susceptibility was assessed according to EUCAST breakpoints (including non-species-specific PK/PD breakpoints, high dosage breakpoints, and breakpoints for the agent without inhibitor, where applicable), except for sulbactam-durlobactam and colistin where FDA breakpoints and ECOFF values were used, respectively. Data are shown where n ≥ 20 isolates were available. Data on susceptibility to colistin are shown in parentheses as colistin is not recommended for monotherapy and is not associated with a clinical monotherapy breakpoint (as per EUCAST v.14.0 guidance).
Refers to susceptibility, or susceptibility with increased exposure for meropenem, meropenem-vaborbactam, aztreonam-avibactam, and cefepime-taniborbactam.
Includes 458 A. baumannii complex isolates.
Sulbactam-durlobactam-resistant isolates were included irrespective of the number of isolates due to the scarcity of published data.
Fig 1.
Cumulative MIC distributions of cefiderocol, BLBLI combinations, and colistin against (A) meropenem-resistant P. aeruginosa (n = 139) and (B) meropenem-resistant Acinetobacter spp. (n = 227). Antimicrobials were tested against P. aeruginosa and/or Acinetobacter spp. based on expected use in a real-world setting. Data on susceptibility to colistin are shown as dashed lines as colistin is not recommended for monotherapy and is not associated with a clinical monotherapy breakpoint (as per EUCAST v.14.0 guidance). Resistance to meropenem was defined using a breakpoint of MIC >8 mg/L, relating to high-dose, extended-infusion (2 g, 3-h infusion) meropenem (as per EUCAST v14.0 guidance). ATM-AVI, aztreonam-avibactam; BLBLI, β-lactam/β-lactamase inhibitor; CST, colistin; C-T, ceftolozane-tazobactam; CZA, ceftazidime-avibactam; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FDC, cefiderocol; FEP-TAN, cefepime-taniborbactam; I-R, imipenem-relebactam; MVB, meropenem-vaborbactam; SUL-DUR, sulbactam-durlobactam.
Susceptibility to cefiderocol remained high against P. aeruginosa isolates resistant to BLBLI combinations, ranging from 93.6% against ceftazidime-avibactam-resistant isolates to 98.1% against imipenem-relebactam-resistant isolates (Table 2). On the other hand, BLBLI combinations did not show high levels of activity against BLBLI combination-resistant isolates, with the highest susceptibility being 71.8% for ceftolozane-tazobactam against aztreonam-avibactam-resistant isolates. Colistin showed 100% susceptibility against BLBLI combination-resistant phenotypes. The activity of antimicrobials against cefiderocol-resistant P. aeruginosa was not analyzed due to the low number of isolates (n = 10).
Against P. aeruginosa isolates that were resistant to both meropenem and ceftazidime-avibactam, susceptibility to cefiderocol was 96.7%, which was higher than any BLBLI combination (≤45.0% susceptibility) (Table 2). Similarly, against meropenem- and ceftolozane-tazobactam-resistant isolates, susceptibility to cefiderocol was higher than all tested BLBLI combinations (98.4% vs <55% susceptibility, respectively). Susceptibility rates for antibiotics against antibiotic-resistant P. aeruginosa using CLSI breakpoints are reported in Table S2.
In total, 45.3% (227/501) Acinetobacter spp. isolates were classified as meropenem resistant (MIC >8 mg/L) (Table 2; see Fig. 1B for MIC distributions). Susceptibility of meropenem-resistant Acinetobacter spp. was 85.0% to cefiderocol [European Committee on Antimicrobial Susceptibility Testing (EUCAST) non-species-specific pharmacokinetic/pharmacodynamic (PK/PD) breakpoint] and 93.8% to sulbactam-durlobactam (FDA breakpoint) (Table 2) (37, 38). The MIC90 was lower for sulbactam-durlobactam (4 mg/L) than cefiderocol (8 mg/L). Cefiderocol susceptibility against sulbactam-durlobactam-resistant isolates (n = 15) was 13.3%, while sulbactam-durlobactam susceptibility against cefiderocol-resistant isolates (n = 38) was 65.8%. Colistin showed high susceptibility (>92.1%) against all resistant phenotypes. Antimicrobial susceptibility rates in antibiotic-resistant Acinetobacter spp. using CLSI or FDA breakpoints are reported in Table S2.
β-Lactamase genes in meropenem-resistant pathogens
Of the 139 meropenem-resistant P. aeruginosa isolates, 2.9% (4/139) were cefiderocol resistant and were analyzed by whole-genome sequencing (WGS), while the remaining isolates were analyzed by PCR for the presence of specific β-lactamase genes. Antimicrobial susceptibility rates in meropenem-resistant P. aeruginosa according to β-lactamase genes identified are reported in Table S5A. The most common β-lactamase genes identified in isolates were MBLs [21.6% (30/139)], including 27 isolates with only MBL genes and 3 isolates that each co-harbored 1 KPC, extended-spectrum β-lactamases (ESBL), or acquired OXA gene (Table 3). This trend was similar across all European countries (Table S6A). A small proportion of meropenem-resistant P. aeruginosa isolates harbored only ESBL genes [5.8% (8/139)] (Table 3). The most common MBL gene in meropenem-resistant P. aeruginosa isolates was VIM [17.3% (24/139), including blaVIM-2 (15/139) and blaVIM-1 (7/139)], while 2.9% (4/139) of the isolates harbored IMP (blaIMP-13: 3/139) and 1.4% (2/139) harbored NDM (blaNDM-1) genes (Table S6A). Overall, 5.0% (7/139) of isolates harbored GES genes (blaGES-5: 6/139), 1.4% (2/139) harbored Vietnamese extended-spectrum β-lactamase (VEB) genes (blaVEB-9), and 0.7% (1/139) harbored blaKPC-2 or blaOXA-10 (Table S6A).
TABLE 3.
| Isolate | β-Lactamase group | Total | |||||
|---|---|---|---|---|---|---|---|
| MBL | MBL + KPC | MBL + ESBL | MBL + OXA | ESBL | Negative | ||
| P. aeruginosa | 27 | 1 | 1 | 1 | 8 | 101 | 139 |
ESBL, extended-spectrum β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; MBL, metallo-β-lactamase; OXA, oxacillinase; PCR, polymerase chain reaction; WGS, whole-genome sequencing.
Data were generated by WGS if meropenem-resistant isolates were resistant to cefiderocol (4/139) or PCR if isolates were susceptible to cefiderocol (135/139). Isolates are grouped as “negative” if only intrinsic β-lactamase genes were present.
Of the 227 meropenem-resistant Acinetobacter spp. isolates, 16.7% (38/227) were cefiderocol resistant and were analyzed by WGS, while the remaining isolates were analyzed by PCR for the presence of specific β-lactamase genes. Antimicrobial susceptibility rates in meropenem-resistant Acinetobacter spp. according to β-lactamase genes identified are reported in Table S5B. The majority of meropenem-resistant Acinetobacter spp. isolates harbored acquired OXA genes [94.7% (215/227)], including 202 isolates with only acquired OXA genes, 9 that co-harbored MBL genes, and 4 that co-harbored ESBL genes (Table 4). This trend was similar across all European countries (Table S6B). A low proportion of isolates harbored only MBL [1.3% (3/227)] genes or co-harbored ESBL and KPC genes [0.4% (1/227)] (Table 4). Overall, 85.5% (194/227) of meropenem-resistant Acinetobacter spp. isolates harbored blaOXA-23 or genes of the OXA-23 group, 14.5% (33/227) harbored blaOXA-24 or genes of the OXA-24 group (including 16/227 isolates that co-harbored blaOXA-23-24), and 3.1% (7/227) harbored blaOXA-72; 5.3% (12/227) of isolates harbored blaNDM-1, and 0.4% (1/227) harbored blaKPC-3 (Table S6B).
TABLE 4.
Acquired β-lactamase genes identified in meropenem-resistant Acinetobacter spp. isolates (n = 227)a,b
| Isolate | β-Lactamase group | Total | |||||
|---|---|---|---|---|---|---|---|
| OXA | OXA + MBL | OXA + ESBL | MBL | ESBL + KPC | Negative | ||
| A. baumannii | 156 | 8 | 3 | 3 | 1 | 6 | 177 |
| A. baumannii complex | 32 | – | – | – | – | 2 | 34 |
| A. bereziniae | 1 | – | – | – | – | – | 1 |
| A. nosocomialis | 1 | – | – | – | – | – | 1 |
| Other Acinetobacter spp. | 12 | 1 | 1 | – | – | – | 14 |
| Total | 202 | 9 | 4 | 3 | 1 | 8 | 227 |
ESBL, extended-spectrum β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; MBL, metallo-β-lactamase; OXA, oxacillinase; PCR, polymerase chain reaction; WGS, whole-genome sequencing.
Data were generated by WGS if meropenem-resistant isolates were resistant to cefiderocol (174/227, including 1 A. baumannii complex and 4 other Acinetobacter spp.) or PCR if isolates were susceptible to cefiderocol (53/227, including 33 A. baumannii complex and 10 other Acinetobacter spp.). Isolates are grouped as “negative” if only intrinsic β-lactamase genes were present (n = 1) or if no screened β-lactamase gene was present (n = 7).
β-Lactamase genes and other potential resistance mechanisms identified in cefiderocol-resistant pathogens
In total, 10 P. aeruginosa and 38 Acinetobacter spp. (37 A. baumannii; 1 Acinetobacter calcoaceticus) isolates were cefiderocol resistant and were analyzed by WGS.
Four cefiderocol-resistant P. aeruginosa isolates were of a sequence type (ST) previously reported (ST235, ST298, ST708, and ST773), while six novel STs were identified (ST4283, ST4290, ST4291, ST4304, ST4305, and ST4306) for isolates collected in France, Germany, Italy, Spain, and the United Kingdom (Table 5). Only one isolate harbored an acquired β-lactamase gene (blaNDM-1). All cefiderocol-resistant isolates had either mutations in piuA and pirA-like genes encoding siderophore uptake receptors or these genes were not detected; the majority also had piuC (9/10) and pvdS (6/10) alterations. Only 20% (2/10) had ftsl and oprD mutations.
TABLE 5.
β-Lactamase genes and other potential resistance mechanisms identified in cefiderocol-resistant P. aeruginosa isolates (n = 10)a,b
| Isolate | Country | FDC MIC (mg/L) | ST | Potential resistance mechanism(s) identified | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactamasec | ftsl | oprD | piuA | piuC | pirA-like | pvdS | ||||
| 1 | France | 8 | 4291d | OXA-396; PDC-543 | WT | No gross disruption | Gene not found | Gene not found | A370T | WT |
| 2 | France | 4 | 773 | OXA-395; PDC-16; NDM-1 | WT | No gross disruption | Q34H | R201H | A370T | V180L; H182N |
| 3 | France | 8 | 235 | OXA-19; OXA-488; PDC-35 | WT | No gross disruption | piuD | R201H | S20N; T235I; A370T |
V180L; H182N |
| 4 | Germany | 4 | 298 | OXA-848; PDC-219 | A244T | Gross disruption | piuD | R201H | Gross disruption | V26A |
| 5 | Germany | 8 | 4290d | OXA-1124; PDC-86 | WT | No gross disruption | Q34H | T10S; R201H |
A13V; A370T | V180L; H182N |
| 6 | Italy | 8 | 708 | OXA-50; PDC-446 | WT | No gross disruption | Q34H | WT | R7H; A370T; Q620K |
V180L; T181A; H182N |
| 7 | Italy | 16 | 4305d | OXA-488; PDC-337 | WT | No gross disruption | Q34H | R201H | A370T | WT |
| 8 | Spain | 4 | 4304d | OXA-1022; PDC-46 | V137M | Gross disruption | Q34H | R201H | Y2S; A370T | WT |
| 9 | Spain | 4 | 4306d | OXA-1188; PDC-5 | WT | No gross disruption | piuD | V104I | A370T | V180L; T181A; H182N; R186P |
| 10 | UK | 4 | 4283d | OXA-1135; PDC-63 | WT | No gross disruption | Gene not found | D49E; E125A; R198Q; V203I | T144S; A208T; A370T; T480A | WT |
FDC, cefiderocol; MBL, metallo-β-lactamase; NDM, New Delhi MBL; OXA, oxacillinase; PDC, Pseudomonas-derived cephalosporinase; ST, sequence type; UK, United Kingdom; WT, wild type.
Data were generated by whole-genome sequencing. A gene was considered to have a gross disruption if the coding sequence carried a nonsense mutation, frameshift, indels of >20 codons, or ablation of the canonical stop or start codons without a replacement immediately adjacent and in-frame. Genes were listed to be not found if a BLAST search with the reference gene yielded no hit with E-value <1E-25. Non-β-lactamase genes that were either found to have gross disruptions or mutations or were not found are shown in gray.
Data shown are a curated summary.
Indicates a novel ST.
Half (19/38) of the cefiderocol-resistant Acinetobacter spp. isolates were found to be ST2, collected in all six European countries, while the other half consisted of isolates of other STs, including two novel STs from France and Germany (Table 6). A total of 84.2% (32/38) of isolates harbored acquired β-lactamase genes. Of the ST2 isolates, 73.7% (14/19) harbored blaOXA-23, 26.3% (5/19) harbored blaOXA-72, and 5.3% (1/19) harbored blaNDM-1 or the blaPER-7 carbapenemase gene. All ST2 isolates had piuA mutations and 42.1% (8/19) had ftsl mutations, but no piuC, pirA-like, or carO gene mutations were observed. Of the non-ST2 isolates, 52.6% (10/19) harbored blaOXA-23 and 57.9% (11/19) harbored blaNDM-1. All non-ST2 isolates had either piuA mutations or this gene was not detected; 57.9% (11/19) had ftsI mutations, and 47.4% (9/19) had pirA-like gene mutations, or this gene was not detected, but no carO mutations were observed.
TABLE 6.
β-Lactamase genes and other potential resistance mechanisms identified in cefiderocol-resistant Acinetobacter spp.a isolates (n = 38)b,c
| Isolate | Country | FDC MIC (mg/L) | ST | Potential resistance mechanism(s) identified | |||||
|---|---|---|---|---|---|---|---|---|---|
| β-Lactamased | ftsI | carO | piuA | piuC | pirA-like | ||||
| 1 | Austria | 8 | 2 | ADC-74; OXA-66; OXA-72; NDM-1 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 2 | Austria | 4 | 2 | ADC-73; OXA-66; OXA-23 | A515V | No gross disruption | Gross disruption | WT | WT |
| 3 | France | 8 | 2 | ADC-85; OXA-66; OXA-23 | A515V | No gross disruption | Gross disruption | WT | WT |
| 4 | France | 8 | 2 | ADC-73; OXA-66; OXA-23 | A515V | No gross disruption | Gross disruption | WT | WT |
| 5 | France | 4 | 2 | ADC-73; OXA-66; OXA-23 | A515V | No gross disruption | Gross disruption | WT | WT |
| 6 | France | >32 | 1 | ADC-191; OXA-69; NDM-1; OXA-23 | L480I; T511S | No gross disruption | Gross disruption | WT | Gross disruption |
| 7 | France | 32 | 1 | ADC-191; OXA-69; NDM-1; OXA-23 | L480I; T511S | No gross disruption | Gross disruption | WT | Y479N; K543R |
| 8 | France | 16 | 85 | ADC-165; OXA-94; NDM-1 | WT | No gross disruption | Gross disruption (~56% ident to reference) |
WT | K475R; H566N |
| 9 | France | >32 | 2163e | ADC-2; OXA-132 | A512T | No gross disruption | I10F; N489D | WT | H566N |
| 10 | France | 8 | 1 | ADC-204; OXA-69; TEM-Trunc; OXA-23 | WT | No gross disruption | L76F; N489D | WT | Y479N; K543R |
| 11 | France | 32 | 85 | ADC-80; OXA-94; NDM-1 | WT | No gross disruption | Gross disruption (~56% ident to reference) |
WT | K475R; H566N |
| 12 | France | 4 | 203 | ADC-Trunc; OXA-65 | WT | No gross disruption | S10T; N19D; R42Q; L52I; N58T; A63S; T66Q; H67Q; Q90N; L119V; E125D; I164V; E185D; N206Q; S208E |
WT | T340I; H566N |
| 13 | Germany | >32 | 600 | ADC-73; OXA-66; TEM-1D; OXA-23 | A515V | No gross disruption | Gross disruption | WT | WT |
| 14 | Germany | >32 | 2 | ADC-30; OXA-66; PER-7; OXA-23 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 15 | Germany | 16 | 85 | ADC-176; OXA-94; NDM-1 |
WT | No gross disruption | Gross disruption (and ~56% ident to reference) |
WT | K475R; H566N |
| 16 | Germany | 4 | 636 | ADC-74; OXA-66; OXA-72 | WT | No gross disruption | Gross disruption | WT | WT |
| 17 | Germany | >32 | 2262e | ADC-11; OXA-66; OXA-72; PER-1 |
WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 18 | Italy | 16 | 2 | ADC-33; OXA-82; OXA-23 | H370Y | Gross disruption | G216V; N489D; K658R | WT | WT |
| 19 | Italy | 4 | 2 | ADC-73; OXA-66; OXA-23 | A515V | No gross disruption | G216V; N489D; K658R | WT | WT |
| 20 | Italy | 4 | 2 | ADC-33; OXA-82; OXA-23 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 21 | Italy | 16 | 2 | ADC-33; OXA-82; OXA-23 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 22 | Italy | 4 | 2 | ADC-33; OXA-82; OXA-23 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 23 | Italy | >32 | 2 | ADC-30; OXA-66; OXA-72 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 24 | Italy | 16 | 2 | ADC-30; OXA-66; OXA-72 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 25 | Italy | >32 | 2 | ADC-30; OXA-66; OXA-72 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 26 | Italy | >32 | 2 | ADC-30; OXA-66; OXA-72 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 27 | Italy | 16 | 600 | ADC-73; OXA-66; TEM-1D; NDM-1; OXA-23 |
A515V | No gross disruption | Gross disruption | WT | WT |
| 28 | Italy | 16 | 600 | ADC-73; OXA-66; TEM-1D; NDM-1; OXA-23 |
A515V | No gross disruption | Gross disruption | WT | WT |
| 29 | Italy | 16 | 600 | ADC-73; OXA-66; TEM-1D; NDM-1; OXA-23 |
A515V | No gross disruption | Gross disruption | WT | WT |
| 30 | Italy | 8 | 600 | ADC-73; OXA-66; TEM-1D; NDM-1; OXA-23 |
A515V | No gross disruption | Gross disruption | WT | WT |
| 31 | Italy | 8 | 600 | ADC-Trunc; OXA-66; TEM-1D; NDM-1; OXA-23 |
A515V | No gross disruption | Gross disruption | WT | WT |
| 32 | Italy | 16 | 600 | ADC-73; OXA-66; TEM-1D; NDM-1; OXA-23 |
A515V | No gross disruption | Gross disruption | WT | WT |
| 33 | Italy | 4 | 2 | ADC-33; OXA-82; OXA-23 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 34 | Italy | 4 | 2 | ADC-33; OXA-82; OXA-23 | WT | No gross disruption | G216V; N489D; K658R | WT | WT |
| 35 | Spain | 4 | 79 | ADC-1; OXA-65 | WT | No gross disruption | Gene not found | Gene not found | WT |
| 36 | Spain | >32 | 2 | ADC-30; OXA-66; OXA-23 | K235N | No gross disruption | G216V; N489D; K658R | WT | WT |
| 37f | UK | 8 | 432 | ADC-295; OXA-1189 | V72I; E115A; T179M; V343I; A345S; Q405E; A435V; E471Q; A483P; P604S; E605V | No gross disruption | Gross disruption | Gene not found | Gene not found |
| 38 | UK | >32 | 2 | ADC-73; OXA-66; TEM-1D; OXA-23 |
A515V | No gross disruption | G216V; N489D; K658R | WT | WT |
Isolates shown are A. baumannii unless otherwise indicated.
ADC, Acinetobacter-derived cephalosporinase; FDC, cefiderocol; MBL, metallo-β-lactamase; NDM, New Delhi MBL; OXA, oxacillinase; ST, sequence type; Trunc, truncated; UK, United Kingdom; WT, wild type.
Data were generated by whole-genome sequencing. A gene was considered to have a gross disruption if the coding sequence carried a nonsense mutation, frameshift, indels of >20 codons, or ablation of the canonical stop or start codons without a replacement immediately adjacent and in-frame. Genes were listed to be not found if a BLAST search with the reference gene yielded no hit with E-value <1E-25. Non-β-lactamase genes that were either found to have gross disruptions or mutations or were not found are shown in gray.
Data shown are a curated summary.
Indicates a novel ST.
A. calcoaceticus.
DISCUSSION
This study provides additional data on the in vitro susceptibilities of cefiderocol and BLBLI combinations, including those still in development, against a large collection of European isolates of glucose non-fermenting Gram-negative bacteria. The data collected in this study are from a greater number of sites per European country compared with the longitudinal SENTRY and SIDERO surveillance programs, which are more geographically spread (35, 36).
The susceptibility rate for cefiderocol was higher than BLBLI combinations (including those still in development, such as aztreonam-avibactam and cefepime-taniborbactam) against P. aeruginosa overall (98.9% vs 83.3%–91.4%, respectively) and meropenem-resistant isolates (97.8% vs ≤59.7%). These observations are consistent with similar previous in vitro studies (5, 26, 39–45). It is important to note that meropenem-resistant P. aeruginosa isolates in this study were defined according to the EUCAST MIC resistance breakpoint for high-dose (2 g), extended (3-h)-infusion meropenem (>8 mg/L), to represent isolates that are meropenem resistant even when treated with the highest meropenem dose available to patients. As some previous studies have defined meropenem-resistant/non-susceptible P. aeruginosa according to the EUCAST MIC resistance breakpoint for standard-dose meropenem (>2 mg/L), susceptibility rates for BLBLI combinations tested in previous studies may be higher (39, 43).
Of the β-lactamase genes observed in meropenem-resistant P. aeruginosa isolates, most were MBLs (most commonly VIM), against which cefiderocol retains high activity, in contrast to most BLBLI combinations (46). Low frequencies (≤5%) of IMP, NDM (blaNDM-1), and GES were observed, lower than previously published data (6). The 71.2% of meropenem-resistant P. aeruginosa isolates which did not harbor β-lactamase genes of interest likely exhibited non-β-lactamase mechanisms of resistance, such as increased expression of efflux systems, chromosomal cephalosporinase activity, or reduced porin expression (3–6, 47).
Susceptibility to cefiderocol was higher than to BLBLI combinations against BLBLI combination-resistant P. aeruginosa (93.6%–98.1% vs 12.2%–71.8%, respectively). Similarly, susceptibility to cefiderocol was high against meropenem-resistant P. aeruginosa resistant to ceftazidime-avibactam or ceftolozane-tazobactam (≥96.7%), while ceftolozane-tazobactam and ceftazidime-avibactam both had poorer activity (≤30.6%). This is indicative of a low degree of cross resistance between cefiderocol and BLBLI combinations and a particularly high degree of cross resistance between ceftolozane-tazobactam and ceftazidime-avibactam. Previous studies have also shown cefiderocol to have much higher activity than ceftazidime-avibactam or ceftolozane-tazobactam against ceftazidime-avibactam- or ceftolozane-tazobactam-resistant P. aeruginosa (6, 48, 49). Given that 11%–23% of P. aeruginosa isolates show resistance to ceftazidime-avibactam or ceftolozane-tazobactam, cefiderocol would be the preferred agent over BLBLI combinations for treatment of infections caused by such isolates. Even aztreonam-avibactam and cefepime-taniborbactam, which are still in development, showed lower susceptibility (≤54.8%) compared with cefiderocol (≥96.7%) against meropenem-resistant P. aeruginosa resistant to ceftazidime-avibactam or ceftolozane-tazobactam. Lower activity of cefepime-taniborbactam was previously demonstrated against ceftolozane-tazobactam- and ceftazidime-avibactam-resistant/-non-susceptible P. aeruginosa, as taniborbactam does not fully restore cefepime activity in some BLBLI combination-resistant P. aeruginosa isolates, such as IMP producers (Table S5) (39, 50). Further, studies on the in vitro activity of aztreonam-avibactam showed poor activity against P. aeruginosa overall (44, 45).
Cefiderocol-resistant P. aeruginosa isolates were collected across Europe in this study. Although isolates were not collected under a surveillance program, the diverse range of STs indicates that cefiderocol resistance in P. aeruginosa arises from specific clones and is not due to clonal expansion. Although there were no clear patterns of resistance mechanisms in cefiderocol-resistant P. aeruginosa isolates in this study, previous observations have noted the common presence of mutations in genes encoding the PiuA and PirA receptors required for the uptake of siderophore conjugates (51–54) and the role of mutations in the ftsI gene encoding PBP3 (50). Further investigations are required to confirm whether mutations in piuA, pirA, and ftsI impact drug resistance or are natural polymorphisms.
Acinetobacter spp. are particularly difficult to treat due to the prevalence of antimicrobial resistance (2). However, both cefiderocol and sulbactam-durlobactam demonstrated good in vitro activity against Acinetobacter spp. overall (>90% susceptibility) and meropenem-resistant isolates (85.0% and 93.8% susceptibility, respectively), in agreement with previous studies (5, 55, 56). Against Acinetobacter spp. overall, cefiderocol demonstrated a lower MIC90 than sulbactam-durlobactam. To the authors’ knowledge, this is the first published study directly comparing the in vitro activity of cefiderocol and sulbactam-durlobactam, which was recently approved by the United States Food and Drug Administration for the treatment of hospital-acquired/ventilator-associated bacterial pneumonia caused by susceptible A. baumannii complex in adults (57, 58). While susceptibility to sulbactam-durlobactam was higher than to cefiderocol against meropenem-resistant Acinetobacter spp., the FDA breakpoint for sulbactam-durlobactam (≤4 mg/L) was used, as EUCAST breakpoints are not available (37). Had the CLSI breakpoint been used in place of the EUCAST non-species-specific PK/PD breakpoint for cefiderocol (≤4 mg/L vs ≤2 mg/L), cefiderocol susceptibility against meropenem-resistant Acinetobacter spp. would be more comparable to sulbactam-durlobactam (88.5%) (Table S2) (59).
The majority of these meropenem-resistant Acinetobacter spp. isolates harbored blaOXA-23 (85.5%), which has been recognized as the most prevalent carbapenem-hydrolyzing class D β-lactamase in CRAB isolates (60–62), although there were low numbers of isolates with MBL genes; 5.3% harbored blaNDM-1 and none harbored blaVIM.
Cefiderocol-resistant Acinetobacter spp. accounted for 7.6% of Acinetobacter spp. collected in this study, against which sulbactam-durlobactam demonstrated good in vitro activity. Although some cefiderocol-resistant isolates had similar genotypic and phenotypic data, which may suggest clonality, isolates were not collected under a surveillance program and numbers were low. Mutations in piuA were found in 97.4% of isolates, suggesting that piuA may be the major iron-regulated outer membrane protein involved in the uptake of cefiderocol in Acinetobacter spp. Half of all cefiderocol-resistant Acinetobacter spp. isolates had mutations in the ftsI gene encoding PBP3, which cefiderocol is known to inhibit (52). The retained activity of sulbactam-durlobactam against cefiderocol-resistant Acinetobacter spp. suggests that there does not appear to be any cross resistance due to PBP3 target-site mutations. This was unexpected, as sulbactam also inhibits PBP3, and sulbactam-durlobactam resistance has previously been attributed to PBP3 mutations (32, 63); in addition, Acinetobacter spp. resistant to sulbactam-durlobactam (3.0% of isolates) also had low susceptibility to cefiderocol (13.3%). It may be a concern that sulbactam-durlobactam resistance is being observed in Europe this early in the use of this treatment, possibly as a consequence of prior exposure to ampicillin-sulbactam. However, this study was not designed to comprehensively investigate mechanisms of resistance in non-fermenter isolates.
The data of this study do provide insights into the therapeutic options for infections due to P. aeruginosa and Acinetobacter spp. with resistant phenotypes. The low levels of cross resistance observed between cefiderocol and any BLBLI combination, with the exception of sulbactam-durlobactam, support the concept that cefiderocol should be tested at the same time as these BLBLI combinations to allow clinicians to choose effective treatment(s) for non-fermenter infections from all available options. It is particularly important that all effective treatment(s) are identified and considered, as current treatment options are limited (33, 34). Importantly, the high cross resistance observed between ceftazidime-avibactam and ceftolozane-tazobactam in this and other studies suggests that cycling between these treatments to treat infections due to P. aeruginosa is unlikely to be an appropriate option (64–66). The susceptibilities of aztreonam-avibactam and cefepime-taniborbactam against P. aeruginosa resistant to meropenem and both meropenem and BLBLI combinations were low, which also suggests that these will not be good treatment options for infections due to P. aeruginosa.
Although susceptibility to colistin was high against meropenem-resistant P. aeruginosa and Acinetobacter spp. isolates, colistin is known to have concerningly high rates of nephrotoxicity and poor tissue penetration, particularly in the lungs (67–69), and the majority of non-fermenter isolates in this study were from respiratory tract infections. Colistin is also not recommended by EUCAST for monotherapy and is not associated with a clinical monotherapy breakpoint (70).
There are several limitations to this study. Isolates were only collected from six European countries, and up to 35 non-fermenter isolates per participating site were included. Hence, there were low numbers of some isolates resistant to at least one antimicrobial, particularly cefiderocol. Ceftazidime-avibactam was tested using a validated commercial method, while other antimicrobials were tested using custom plates. A selection of potential mechanisms of resistance was only screened for in meropenem- and cefiderocol-resistant isolates, using different methodologies and screening panels, and excluding analysis of expression levels of genes or other potential mechanisms of BLBLI combination resistance, such as PBP1 and PBP2. Therefore, robust interpretations of resistance mechanisms and any cross resistance could not be made. The STs of cefiderocol-susceptible isolates were not identified, so clonal expansion of isolates with resistant phenotypes was not determined; however, these non-surveillance data would not have accurately reflected clonal epidemiology in Europe. Lastly, in vitro data cannot replace clinical studies in patients, and in vitro activity may not reflect in vivo efficacy of a therapy in clinical practice.
Conclusions
These results confirm the high levels of in vitro activity of cefiderocol against Gram-negative P. aeruginosa and Acinetobacter spp. isolates from Europe, including both meropenem-resistant isolates and those resistant to recent BLBLI combinations commonly used in first-line treatment of CR infections. Cefiderocol often had high in vitro activity where the majority of BLBLI combinations did not, and there was no apparent cross resistance between cefiderocol and BLBLI combinations, with the exception of sulbactam-durlobactam.
MATERIALS AND METHODS
Clinical isolates
Between 01 January and 31 December 2020, Gram-negative clinical isolates from hospitalized inpatients were collected at 49 sites across Austria, France, Germany, Italy, Spain, and the United Kingdom (see Table S7 for details of participating centers). Each site was requested to collect 20 P. aeruginosa and 15 A. baumannii (Enterobacterales isolates were also collected as part of the overall study, for which the methods and results are reported elsewhere). Isolates included those from all infection sources, with the exception of the urinary tract. Only one isolate of the same genus and species was allowed per patient. Matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry was used for species identification at International Health Management Associates (IHMA) Europe Sàrl (Monthey, Switzerland).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed on all collected isolates at IHMA Europe Sàrl. Isolates were stored at −70°C before testing by broth microdilution for the determination of MICs. Antimicrobials tested were cefiderocol, meropenem, ceftazidime-avibactam, ceftolozane-tazobactam, meropenem-vaborbactam, imipenem-relebactam, aztreonam-avibactam, cefepime-taniborbactam, and colistin against P. aeruginosa, and cefiderocol, meropenem, sulbactam-durlobactam, and colistin against Acinetobacter spp. (see Table S8 for suppliers of agents).
International Organization for Standardization 20776-1 susceptibility testing standards and EUCAST guidance were followed for the preparation of antimicrobials for testing and MIC determinations (71, 72); tryptic soy agar plates containing 5% sheep blood were sourced from Liofilchem (Roseto degli Abruzzi, Italy; product code: 11037), cation-adjusted Mueller-Hinton broth was sourced from Becton Dickinson (Franklin Lakes, NJ, USA; product code: 212322), and iron-depleted cation-adjusted Mueller-Hinton broth (used for cefiderocol testing) was prepared by IHMA Europe Sàrl. This excludes ceftazidime-avibactam, for which MIC values were only available when Sensititre freeze-dried panels (Thermo Fisher Scientific Inc., Waltham, MA, USA) were used in the preparation of ceftazidime-avibactam for testing. All antibiotics were tested daily using the quality control strains P. aeruginosa ATCC 27853 (as recommended by EUCAST and CLSI, and in line with guidance from CLSI) and A. baumannii ATCC 13304 (in line with guidance from CLSI, where a same-species quality control strain was not recommended by EUCAST) (59, 73). The MIC values for each tested antibiotic were manually read as the lowest concentration inhibiting visible growth. For cefiderocol and meropenem, MIC values were determined more than once; a third MIC determination was carried out if MIC values differed by >1 dilution, and the geometric mean was reported.
Analysis
Antimicrobial susceptibility results were interpreted in accordance with EUCAST clinical breakpoints (v.14.0, 2024) (37) (see Table S9 for breakpoints used).
Meropenem resistance was defined using a breakpoint of MIC >8 mg/L, relating to high-dose, extended-infusion (2 g, 3-h infusion) meropenem; similarly, isolates with a meropenem MIC >8 mg/L when tested with a fixed vaborbactam concentration of 8 mg/L were considered resistant to meropenem-vaborbactam. Aztreonam-avibactam, cefepime-taniborbactam, and sulbactam-durlobactam do not currently have approved EUCAST MIC breakpoints; nor does cefiderocol for Acinetobacter spp. For aztreonam-avibactam and cefepime-taniborbactam, EUCAST breakpoints for high-dose aztreonam and high-dose cefepime alone were used. For Acinetobacter spp., the non-species-specific PK/PD breakpoint (37) was used for cefiderocol and the FDA breakpoint (38) was used for sulbactam-durlobactam. For colistin, EUCAST ECOFF values were used.
Identification of β-lactamase genes in meropenem-resistant isolates
Isolates with a meropenem MIC >8 mg/L and a cefiderocol MIC ≤2 mg/L were analyzed by PCR (performed by IHMA Europe Sàrl) to identify the presence of β-lactamase genes that may confer meropenem resistance (see Table S10 for genes and primers used). Data on β-lactamase genes in isolates that were meropenem resistant (meropenem MIC >8 mg/L) and cefiderocol resistant (MIC >2 mg/L) were generated by WGS (see below).
DNA extraction was performed from a single colony obtained from a fresh tryptic soy blood agar culture for each isolate, using the QIAGEN TissueLyser II instrument (Hilden, Germany) as per manufacturer instructions. Preparations then underwent PCR amplification and sequencing to screen for the presence of genes encoding clinically relevant β-lactamases: ESBLs (blaSHV, blaTEM, blaCTX-M, blaVEB, blaPER, blaGES), AmpCs [blaACC, blaCMY I/MOX, blaCMY II, blaDHA, blaFOX, blaACT-MIR, blaPDC (in P. aeruginosa only)], and carbapenemases (blaKPC, blaOXA, blaNDM, blaIMP, blaVIM, blaSPM, blaGIM, blaGES). Amplicons were sequenced by Fasteris (Geneva, Switzerland) and then analyzed using SeqScape Software 3 (Thermo Fisher Scientific Inc.; Waltham, MA, USA). Limited sequencing was used to screen blaTEM and blaSHV to identify TEM-type and SHV-type enzymes containing amino acid substitutions common to ESBLs (blaTEM: amino acid 104, 164, 238, 240; blaSHV: amino acid 146, 179, 238, 240) and to screen blaCTX-M (groups 1, 2, 8, 9, and 25) to identify CTX-M-type enzymes containing the D240G amino acid substitution associated with elevated ceftazidime MIC values. Genes encoding SHV-type and TEM-type enzymes were reported as ESBL or original-spectrum β-lactamase genes. The 16S ribosomal DNA for all isolates was also amplified by PCR and sequenced for bacterial identification.
Identification of β-lactamase genes and other potential resistance mechanisms in cefiderocol-resistant isolates
Isolates with a cefiderocol MIC >2 mg/L were analyzed by WGS to identify possible mechanisms of resistance. DNA isolation was performed using the QIAGEN QIAamp DNA Mini kit, and library preparation was performed using the Illumina DNA Prep kit (San Diego, CA, USA) at IHMA, Inc. (Schaumburg, IL, USA). Libraries were then shipped to Azenta (South Plainfield, NJ, USA), where short-read WGS (2 × 150 base pairs; paired-end) was performed on an Illumina HiSeq platform to a 100× depth of coverage. Quality control was performed using the CheckM lineage workflow (74–76) to assure low contamination (≤5%) and completeness of assemblies (≥95%) were achieved. The multilocus sequence typing scheme Pasteur was used to determine relatedness of Acinetobacter spp. isolates.
Genomic assemblies were created using the QIAGEN CLC Genomics workbench (v.21.0.5). In order to identify β-lactamase genes of interest, assemblies were queried using the ResFinder database (77) with coverage and identity thresholds of ≥35% and ≥72%, respectively. Genes identified with <100% identity or coverage were evaluated for a variant by pairwise alignment to a reference sequence using the ResFinder database (77). Variants were defined using the Bacterial Antimicrobial Resistance Reference Gene Database from the National Center for Biotechnology Information (Bioproject 313047).
Non-β-lactamase genes of interest in this study included those encoding PBP3 (ftsI), porins [oprD (P. aeruginosa) and carO (Acinetobacter spp.)], and those related to iron acquisition [pirA-like, piuA, piuC, and pvdS (P. aeruginosa)]. Genes were analyzed by pairwise alignment and classified as wild type if they had 100% amino acid sequence identity to the species-specific reference sequence (Table S11). These genes were also screened for gross disruption vs species-specific reference sequences (Table S11) and were considered to have gross disruption if the coding sequence carried a nonsense mutation, frameshift, indels of >20 codons, or ablation of the canonical start or stop codons without a replacement immediately adjacent and in-frame. Genes were not considered disrupted if there were ablated start or stop codons immediately adjacent to intact, in-frame start or stop codons. Genes were listed to be not found if a BLAST search with the reference gene yielded no hit with E-value <1E−25.
ACKNOWLEDGMENTS
The authors would like to thank all investigators and participating sites for the provision of clinical isolates. The ARTEMIS study was designed by Shionogi B.V., Amsterdam, the Netherlands. Coordination of the ARTEMIS study, including generation of the in vitro MIC data, was carried out by IHMA Europe Sàrl, Monthey, Switzerland, and genetic sequencing of isolates was carried out by IHMA, Inc., Schaumburg, IL, USA, with funding by Shionogi B.V. Editorial assistance in the preparation of this manuscript, under the direction of the authors, was provided by Hannah James, MSc, of Ashfield MedComms, an Inizio Company, funded by Shionogi B.V. and complied with Good Publication Practice (GPP) guidelines (78).
This work was supported by Shionogi B.V., Amsterdam, the Netherlands.
ARTEMIS Study Investigators: Birgit Willinger, David Leyssene, Christian Cattoen, Corentine Alauzet, Pierre Boyer, Véronique Dubois, Katy Jeannot, Stephane Corvec, Jean-Philippe Lavigne, Thomas Guillard, Audrey Merens Gontier, Thierry Naas, Holger Rohde, Stefan Ziesing, Can Imirzalioglu, Klaus-Peter Hunfeld, Jette Jung, Sören Gatermann, Mathias Pletz, Gabriele Bianco, Anna Giammanco, Davide Carcione, Giammarco Raponi, Caterina Matinato, Enea Gino Di Domenico, Paolo Gaibani, Anna Marchese, Fabio Arena, Claudia Niccolai, Stefania Stefani, Cristina Pitart, Jose Luis Barrios, Emilia Cercenado, German Bou, Alicia Beteta Lopez, Rafael Canton, Jose Lopez Hontangas, Irene Gracia-Ahufinger, Antonio Oliver, Lorena Lopez-Cerero, Nieves Larrosa, David Wareham, John Perry, Anna Casey, Jasvir Nahl, Daniel Hughes, Michael Coyne, Michelle Lister, Marie Attwood.
All authors contributed toward data curation and analysis, writing, reviewing, and editing the manuscript and agreed to be accountable for all aspects of the work.
Contributor Information
Anne Santerre Henriksen, Email: anne.henriksen@shionogi.eu.
Rolf Müller, Helmholtz-Institut fur Pharmazeutische Forschung Saarland, Saarbrücken, Germany.
ARTEMIS Study Investigators:
Birgit Willinger, David Leyssene, Christian Cattoen, Corentine Alauzet, Pierre Boyer, Véronique Dubois, Katy Jeannot, Stephane Corvec, Jean-Philippe Lavigne, Thomas Guillard, Audrey Merens Gontier, Thierry Naas, Holger Rohde, Stefan Ziesing, Can Imirzalioglu, Klaus-Peter Hunfeld, Jette Jung, Sören Gatermann, Mathias Pletz, Gabriele Bianco, Anna Giammanco, Davide Carcione, Giammarco Raponi, Caterina Matinato, Enea Gino Di Domenico, Paolo Gaibani, Anna Marchese, Fabio Arena, Claudia Niccolai, Stefania Stefani, Cristina Pitart, Jose Luis Barrios, Emilia Cercenado, German Bou, Alicia Beteta Lopez, Rafael Canton, Jose Lopez Hontangas, Irene Gracia-Ahufinger, Antonio Oliver, Lorena Lopez-Cerero, Nieves Larrosa, David Wareham, John Perry, Anna Casey, Jasvir Nahl, Daniel Hughes, Michael Coyne, Michelle Lister, and Marie Attwood
ETHICS APPROVAL
Ethics approval was not required as all in vitro samples were anonymized.
DATA AVAILABILITY
Whole-genome sequencing variants were defined using the Bacterial Antimicrobial Resistance Reference Gene Database from the National Center for Biotechnology Information (Bioproject number: PRJNA313047). Data are available from Shionogi B.V. upon reasonable request.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03836-23.
Figures S1; Tables S1 to S11.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. European Centre for Disease Prevention and Control . 2022. Antimicrobial resistance in the EU/EEA (EARS-Net) - annual epidemiological report for 2021. Stockholm: ECDC [Google Scholar]
- 2. World Health Organization . 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis.
- 3. Bubonja-Sonje M, Matovina M, Skrobonja I, Bedenic B, Abram M. 2015. Mechanisms of carbapenem resistance in multidrug-resistant clinical isolates of Pseudomonas aeruginosa from a croatian hospital. Microb Drug Resist 21:261–269. doi: 10.1089/mdr.2014.0172 [DOI] [PubMed] [Google Scholar]
- 4. De Rosa A, Mutters NT, Mastroianni CM, Kaiser SJ, Günther F. 2019. Distribution of carbapenem resistance mechanisms in clinical isolates of XDR Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 38:1547–1552. doi: 10.1007/s10096-019-03585-0 [DOI] [PubMed] [Google Scholar]
- 5. Longshaw C, Manissero D, Tsuji M, Echols R, Yamano Y. 2020. In vitro activity of the siderophore cephalosporin, cefiderocol, against molecularly characterized, carbapenem-non-susceptible Gram-negative bacteria from Europe. JAC Antimicrob Resist 2:dlaa060. doi: 10.1093/jacamr/dlaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Torrens G, van der Schalk TE, Cortes-Lara S, Timbermont L, Del Barrio-Tofiño E, Xavier BB, Zamorano L, Lammens C, Ali O, Ruzin A, Goossens H, Kumar-Singh S, Kluytmans J, Paling F, MacLean RC, Köhler T, López-Causapé C, Malhotra-Kumar S, Oliver A, ASPIRE-ICU study team . 2022. Susceptibility profiles and resistance genomics of Pseudomonas aeruginosa isolates from European ICUs participating in the ASPIRE-ICU trial. J Antimicrob Chemother 77:1862–1872. doi: 10.1093/jac/dkac122 [DOI] [PubMed] [Google Scholar]
- 7. Reyes J, Komarow L, Chen L, Ge L, Hanson BM, Cober E, Herc E, Alenazi T, Kaye KS, Garcia-Diaz J, et al. 2023. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 4:e159–e170. doi: 10.1016/S2666-5247(22)00329-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Del Barrio-Tofiño E, López-Causapé C, Oliver A. 2020. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents 56:106196. doi: 10.1016/j.ijantimicag.2020.106196 [DOI] [PubMed] [Google Scholar]
- 9. Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Kostyanev T, Xavier BB, García-Castillo M, Lammens C, Bravo-Ferrer Acosta J, Rodríguez-Baño J, Cantón R, Glupczynski Y, Goossens H, EURECA/WP1B Group . 2021. Phenotypic and molecular characterizations of carbapenem-resistant Acinetobacter baumannii isolates collected within the EURECA study. Int J Antimicrob Agents 57:106345. doi: 10.1016/j.ijantimicag.2021.106345 [DOI] [PubMed] [Google Scholar]
- 11. Gupta N, Angadi K, Jadhav S. 2022. Molecular characterization of carbapenem-resistant Acinetobacter baumannii with special reference to carbapenemases: a systematic review. Infect Drug Resist 15:7631–7650. doi: 10.2147/IDR.S386641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hujer AM, Hujer KM, Leonard DA, Powers RA, Wallar BJ, Mack AR, Taracila MA, Rather PN, Higgins PG, Prati F, Caselli E, Marshall SH, Clarke T, Greco C, Venepally P, Brinkac L, Kreiswirth BN, Fouts DE, Bonomo RA, Antibacterial Resistance Leadership Group (ARLG) . 2021. A comprehensive and contemporary "snapshot" of β-lactamases in carbapenem resistant Acinetobacter baumannii. Diagn Microbiol Infect Dis 99:115242. doi: 10.1016/j.diagmicrobio.2020.115242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ambrosi C, Scribano D, Aleandri M, Zagaglia C, Di Francesco L, Putignani L, Palamara AT. 2017. Acinetobacter baumannii virulence traits: a comparative study of a novel sequence type with other Italian endemic international clones. Front Microbiol 8:1977. doi: 10.3389/fmicb.2017.01977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nowak P, Paluchowska P. 2016. Acinetobacter baumannii: biology and drug resistance - role of carbapenemases. Folia Histochem Cytobiol 54:61–74. doi: 10.5603/FHC.a2016.0009 [DOI] [PubMed] [Google Scholar]
- 15. European Medicines Agency . 2023. Fetcroja summary of product characteristics.
- 16. Ito A, Kohira N, Bouchillon SK, West J, Rittenhouse S, Sader HS, Rhomberg PR, Jones RN, Yoshizawa H, Nakamura R, Tsuji M, Yamano Y. 2016. In vitro antimicrobial activity of S-649266, a catechol-substituted siderophore cephalosporin, when tested against non-fermenting Gram-negative bacteria. J Antimicrob Chemother 71:670–677. doi: 10.1093/jac/dkv402 [DOI] [PubMed] [Google Scholar]
- 17. Ito A, Nishikawa T, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, Tsuji M, Yamano Y. 2016. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:7396–7401. doi: 10.1128/AAC.01405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ito-Horiyama T, Ishii Y, Ito A, Sato T, Nakamura R, Fukuhara N, Tsuji M, Yamano Y, Yamaguchi K, Tateda K. 2016. Stability of novel siderophore cephalosporin S-649266 against clinically relevant carbapenemases. Antimicrob Agents Chemother 60:4384–4386. doi: 10.1128/AAC.03098-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mushtaq S, Sadouki Z, Vickers A, Livermore DM, Woodford N. 2020. In vitro activity of cefiderocol, a siderophore cephalosporin, against multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother 64:e01582-20. doi: 10.1128/AAC.01582-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Canton R, Doi Y, Simner PJ. 2022. Treatment of carbapenem-resistant Pseudomonas aeruginosa infections: a case for cefiderocol. Expert Rev Anti Infect Ther 20:1077–1094. doi: 10.1080/14787210.2022.2071701 [DOI] [PubMed] [Google Scholar]
- 21. Zhanel GG, Golden AR, Zelenitsky S, Wiebe K, Lawrence CK, Adam HJ, Idowu T, Domalaon R, Schweizer F, Zhanel MA, Lagacé-Wiens PRS, Walkty AJ, Noreddin A, Lynch Iii JP, Karlowsky JA. 2019. Cefiderocol: a siderophore cephalosporin with activity against carbapenem-resistant and multidrug-resistant Gram-negative bacilli. Drugs 79:271–289. doi: 10.1007/s40265-019-1055-2 [DOI] [PubMed] [Google Scholar]
- 22. Sader HS, Farrell DJ, Castanheira M, Flamm RK, Jones RN. 2014. Antimicrobial activity of ceftolozane/tazobactam tested against Pseudomonas aeruginosa and Enterobacteriaceae with various resistance patterns isolated in European hospitals (2011-12). J Antimicrob Chemother 69:2713–2722. doi: 10.1093/jac/dku184 [DOI] [PubMed] [Google Scholar]
- 23. Sader HS, Rhomberg PR, Farrell DJ, Jones RN. 2011. Antimicrobial activity of CXA-101, a novel cephalosporin tested in combination with tazobactam against Enterobacteriaceae, Pseudomonas aeruginosa, and Bacteroides fragilis strains having various resistance phenotypes. Antimicrob Agents Chemother 55:2390–2394. doi: 10.1128/AAC.01737-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Farrell DJ, Sader HS, Flamm RK, Jones RN. 2014. Ceftolozane/tazobactam activity tested against Gram-negative bacterial isolates from hospitalised patients with pneumonia in US and European medical centres (2012). J Antimicrob Agents 43:533–539. doi: 10.1016/j.ijantimicag.2014.01.032 [DOI] [PubMed] [Google Scholar]
- 25. Lagacé-Wiens P, Walkty A, Karlowsky JA. 2014. Ceftazidime-avibactam: an evidence-based review of its pharmacology and potential use in the treatment of Gram-negative bacterial infections. Core Evid 9:13–25. doi: 10.2147/CE.S40698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hernández-García M, García-Castillo M, Ruiz-Garbajosa P, Bou G, Siller-Ruiz M, Pitart C, Gracia-Ahufinger I, Mulet X, Pascual Á, Tormo N, Cantón R. 2022. In vitro activity of cefepime-taniborbactam against carbapenemase-producing Enterobacterales and Pseudomonas aeruginosa isolates recovered in Spain. Antimicrob Agents Chemother 66:e0216121. doi: 10.1128/aac.02161-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamrick JC, Docquier J-D, Uehara T, Myers CL, Six DA, Chatwin CL, John KJ, Vernacchio SF, Cusick SM, Trout REL, Pozzi C, De Luca F, Benvenuti M, Mangani S, Liu B, Jackson RW, Moeck G, Xerri L, Burns CJ, Pevear DC, Daigle DM. 2020. VNRX-5133 (Taniborbactam), a broad-spectrum inhibitor of serine- and metallo-β-lactamases, restores activity of cefepime in Enterobacterales and Pseudomonas aeruginosa. Antimicrob Agents Chemother 64:e01963-19. doi: 10.1128/AAC.01963-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wi YM, Greenwood-Quaintance KE, Schuetz AN, Ko KS, Peck KR, Song JH, Patel R. 2018. Activity of ceftolozane-tazobactam against carbapenem-resistant, non-carbapenemase-producing Pseudomonas aeruginosa and associated resistance mechanisms. Antimicrob Agents Chemother 62:e01970-17. doi: 10.1128/AAC.01970-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kazmierczak KM, Tsuji M, Wise MG, Hackel M, Yamano Y, Echols R, Sahm DF. 2019. In vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptible Gram-negative bacilli, including serine carbapenemase- and metallo-β-lactamase-producing isolates (SIDERO-WT-2014 study). Int J Antimicrob Agents 53:177–184. doi: 10.1016/j.ijantimicag.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 30. Mushtaq S, Vickers A, Doumith M, Ellington MJ, Woodford N, Livermore DM. 2021. Activity of β-lactam/taniborbactam (VNRX-5133) combinations against carbapenem-resistant Gram-negative bacteria. J Antimicrob Chemother 76:160–170. doi: 10.1093/jac/dkaa391 [DOI] [PubMed] [Google Scholar]
- 31. McLeod SM, Moussa SH, Hackel MA, Miller AA. 2020. In vitro activity of sulbactam-durlobactam against Acinetobacter baumannii-calcoaceticus complex isolates collected globally in 2016 and 2017. Antimicrob Agents Chemother 64:e02534-19. doi: 10.1128/AAC.02534-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Principe L, Di Bella S, Conti J, Perilli M, Piccirilli A, Mussini C, Decorti G. 2022. Acinetobacter baumannii resistance to sulbactam/durlobactam: a systematic review. Antibiotics 11:1793. doi: 10.3390/antibiotics11121793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paul M, Carrara E, Retamar P, Tängdén T, Bitterman R, Bonomo RA, de Waele J, Daikos GL, Akova M, Harbarth S, Pulcini C, Garnacho-Montero J, Seme K, Tumbarello M, Lindemann PC, Gandra S, Yu Y, Bassetti M, Mouton JW, Tacconelli E, Rodríguez-Baño J. 2022. European society of clinical microbiology and infectious diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European society of intensive care medicine). Clin Microbiol Infect 28:521–547. doi: 10.1016/j.cmi.2021.11.025 [DOI] [PubMed] [Google Scholar]
- 34. Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. 2023. Infectious diseases society of America antimicrobial-resistant treatment guidance: Gram-negative bacterial infections. Clin Infect Dis:ciad428. doi: 10.1093/cid/ciad428 [DOI] [PubMed] [Google Scholar]
- 35. Shortridge D, Streit JM, Mendes R, Castanheira M. 2022. In vitro activity of cefiderocol against U.S. and European Gram-negative clinical isolates collected in 2020 as part of the SENTRY antimicrobial surveillance program. Microbiol Spectr 10:e0271221. doi: 10.1128/spectrum.02712-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takemura M, Wise MG, Hackel MA, Sahm DF, Yamano Y. 2023. In vitro activity of cefiderocol against MBL-producing Gram-negative bacteria collected in North America and Europe in five consecutive annual multinational SIDERO-WT surveillance studies (2014–2019). J Antimicrob Chemother 78:2019–2027. doi: 10.1093/jac/dkad200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. European Committee on Antimicrobial Susceptibility Testing . 2024. Breakpoint tables for interpretation of MICs and zone diameters. Version 14.0
- 38. FDA . Sulbactam and durlobactam injection. Available from: https://www.fda.gov/drugs/development-resources/sulbactam-and-durlobactam-injection. Retrieved 29 Jan 2024.
- 39. Karlowsky JA, Hackel MA, Takemura M, Yamano Y, Echols R, Sahm DF. 2022. In vitro susceptibility of Gram-negative pathogens to cefiderocol in five consecutive annual multinational SIDERO-WT surveillance studies, 2014 to 2019. Antimicrob Agents Chemother 66:e0199021. doi: 10.1128/AAC.01990-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. 2017. In vitro activity of the siderophore cephalosporin, cefiderocol, against a recent collection of clinically relevant Gram-negative bacilli from North America and Europe, including carbapenem-nonsusceptible isolates (SIDERO-WT-2014 study). Antimicrob Agents Chemother 61:e00093-17. doi: 10.1128/AAC.00093-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oueslati S, Bogaerts P, Dortet L, Bernabeu S, Ben Lakhal H, Longshaw C, Glupczynski Y, Naas T. 2022. In vitro activity of cefiderocol and comparators against carbapenem-resistant Gram-negative pathogens from France and Belgium. Antibiotics (Basel) 11:1352. doi: 10.3390/antibiotics11101352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jacobs MR, Abdelhamed AM, Good CE, Rhoads DD, Hujer KM, Hujer AM, Domitrovic TN, Rudin SD, Richter SS, van Duin D, Kreiswirth BN, Greco C, Fouts DE, Bonomo RA. 2019. ARGONAUT-I: activity of cefiderocol (S-649266), a siderophore cephalosporin, against Gram-negative bacteria, including carbapenem-resistant nonfermenters and Enterobacteriaceae with defined extended-spectrum β-lactamases and carbapenemases. Antimicrob Agents Chemother 63:e01801-18. doi: 10.1128/AAC.01801-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karlowsky JA, Lob SH, Akrich B, DeRyke CA, Siddiqui F, Young K, Motyl MR, Hawser SP, Sahm DF. 2023. In vitro activity of imipenem/relebactam against piperacillin/tazobactam-resistant and meropenem-resistant non-morganellaceae Enterobacterales and Pseudomonas aeruginosa collected from patients with lower respiratory tract infections in Western Europe: SMART 2018-20. JAC Antimicrob Resist 5:dlad003. doi: 10.1093/jacamr/dlad003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karlowsky JA, Kazmierczak KM, de Jonge BLM, Hackel MA, Sahm DF, Bradford PA. 2017. In vitro activity of aztreonam-avibactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated by clinical laboratories in 40 countries from 2012 to 2015. Antimicrob Agents Chemother 61:e00472-17. doi: 10.1128/AAC.00472-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biedenbach DJ, Kazmierczak K, Bouchillon SK, Sahm DF, Bradford PA. 2015. In vitro activity of aztreonam-avibactam against a global collection of Gram-negative pathogens from 2012 and 2013. Antimicrob Agents Chemother 59:4239–4248. doi: 10.1128/AAC.00206-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wise MG, Karlowsky JA, Hackel MA, Takemura M, Yamano Y, Echols R, Sahm DF. 2023. In vitro activity of cefiderocol against meropenem-nonsusceptible Gram-negative bacilli with defined β-lactamase carriage: SIDERO-WT surveillance studies, 2014–2019. Microb Drug Resist 29:360–370. doi: 10.1089/mdr.2022.0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hammoudi Halat D, Ayoub Moubareck C. 2022. The intriguing carbapenemases of Pseudomonas aeruginosa: current status, genetic profile, and global epidemiology. Yale J Biol Med 95:507–515. [PMC free article] [PubMed] [Google Scholar]
- 48. Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Martín-Pena ML, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. doi: 10.1093/jac/dkx424 [DOI] [PubMed] [Google Scholar]
- 49. Haidar G, Philips NJ, Shields RK, Snyder D, Cheng S, Potoski BA, Doi Y, Hao B, Press EG, Cooper VS, Clancy CJ, Nguyen MH. 2017. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis 65:110–120. doi: 10.1093/cid/cix182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lasarte-Monterrubio C, Fraile-Ribot PA, Vázquez-Ucha JC, Cabot G, Guijarro-Sánchez P, Alonso-García I, Rumbo-Feal S, Galán-Sánchez F, Beceiro A, Arca-Suárez J, Oliver A, Bou G. 2022. Activity of cefiderocol, imipenem/relebactam, cefepime/taniborbactam and cefepime/zidebactam against ceftolozane/tazobactam- and ceftazidime/avibactam-resistant Pseudomonas aeruginosa. J Antimicrob Chemother 77:2809–2815. doi: 10.1093/jac/dkac241 [DOI] [PubMed] [Google Scholar]
- 51. Moynié L, Luscher A, Rolo D, Pletzer D, Tortajada A, Weingart H, Braun Y, Page MGP, Naismith JH, Köhler T. 2017. Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother 61:e02531-16. doi: 10.1128/AAC.02531-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ito A, Sato T, Ota M, Takemura M, Nishikawa T, Toba S, Kohira N, Miyagawa S, Ishibashi N, Matsumoto S, Nakamura R, Tsuji M, Yamano Y. 2018. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob Agents Chemother 62:e01454-17. doi: 10.1128/AAC.01454-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luscher A, Moynié L, Auguste PS, Bumann D, Mazza L, Pletzer D, Naismith JH, Köhler T. 2018. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugates. Antimicrob Agents Chemother 62:e00097-18. doi: 10.1128/AAC.00097-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luscher A, Gasser V, Bumann D, Mislin GLA, Schalk IJ, Köhler T. 2022. Plant-derived catechols are substrates of TonB-dependent transporters and sensitize Pseudomonas aeruginosa to siderophore-drug conjugates. mBio 13:e0149822. doi: 10.1128/mbio.01498-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Karlowsky JA, Hackel MA, McLeod SM, Miller AA. 2022. In vitro activity of sulbactam-durlobactam against global isolates of Acinetobacter baumannii-calcoaceticus complex collected from 2016 to 2021. Antimicrob Agents Chemother 66:e0078122. doi: 10.1128/aac.00781-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petropoulou D, Siopi M, Vourli S, Pournaras S. 2021. Activity of sulbactam-durlobactam and comparators against a national collection of carbapenem-resistant Acinetobacter baumannii isolates from Greece. Front Cell Infect Microbiol 11:814530. doi: 10.3389/fcimb.2021.814530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. FDA . 2023. FDA approves new treatment for pneumonia caused by certain difficult-to-treat bacteria, on FDA.gov. Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-new-treatment-pneumonia-caused-certain-difficult-treat-bacteria. Retrieved 29 Jan 2024.
- 58. ClinicalTrials.gov . 2023. Study to evaluate the efficacy and safety of intravenous sulbactam-ETX2514 in the treatment of patients with infections caused by Acinetobacter baumannii-calcoaceticus complex (ATTACK), on ClinicalTrials.gov. Available from: https://clinicaltrials.gov/ct2/show/NCT03894046. Retrieved 29 Jan 2024.
- 59. Clinical and Laboratory Standards Institute . 2023. Performance standards for antimicrobial susceptibility testing. 33rd ed. CLSI M100. [Google Scholar]
- 60. D’Onofrio V, Conzemius R, Varda-Brkić D, Bogdan M, Grisold A, Gyssens IC, Bedenić B, Barišić I. 2020. Epidemiology of colistin-resistant, carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Croatia. Infect Genet Evol 81:104263. doi: 10.1016/j.meegid.2020.104263 [DOI] [PubMed] [Google Scholar]
- 61. Karampatakis T, Antachopoulos C, Tsakris A, Roilides EJFM. 2017. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in Greece: an extended review (2000–2015). J Future Microbiol 12:801–815. doi: 10.2217/fmb-2016-0200 [DOI] [PubMed] [Google Scholar]
- 62. Zeka AN, Poirel L, Sipahi OR, Bonnin RA, Arda B, Ozinel M, Ulusoy S, Bor C, Nordmann P. 2014. GES-type and OXA-23 carbapenemase-producing Acinetobacter baumannii in Turkey. J Antimicrob Chemother 69:1145–1146. doi: 10.1093/jac/dkt465 [DOI] [PubMed] [Google Scholar]
- 63. Penwell WF, Shapiro AB, Giacobbe RA, Gu R-F, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BLM, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. doi: 10.1128/AAC.04808-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meschiari M, Orlando G, Kaleci S, Bianco V, Sarti M, Venturelli C, Mussini C. 2021. Combined resistance to ceftolozane-tazobactam and ceftazidime-avibactam in extensively drug-resistant (XDR) and multidrug-resistant (MDR) Pseudomonas aeruginosa: resistance predictors and impact on clinical outcomes besides implications for antimicrobial stewardship programs. Antibiotics (Basel) 10:1224. doi: 10.3390/antibiotics10101224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sader HS, Duncan LR, Doyle TB, Castanheira M. 2021. Antimicrobial activity of ceftazidime/avibactam, ceftolozane/tazobactam and comparator agents against Pseudomonas aeruginosa from cystic fibrosis patients. JAC Antimicrob Resist 3:dlab126. doi: 10.1093/jacamr/dlab126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rubio AM, Kline EG, Jones CE, Chen L, Kreiswirth BN, Nguyen MH, Clancy CJ, Cooper VS, Haidar G, Van Tyne D, Shields RK. 2021. In vitro susceptibility of multidrug-resistant Pseudomonas aeruginosa following treatment-emergent resistance to ceftolozane-tazobactam. Antimicrob Agents Chemother 65:e00084-21. doi: 10.1128/AAC.00084-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eljaaly K, Bidell MR, Gandhi RG, Alshehri S, Enani MA, Al-Jedai A, Lee TC. 2021. Colistin nephrotoxicity: meta-analysis of randomized controlled trials. Open Forum Infect Dis 8:ofab026. doi: 10.1093/ofid/ofab026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Michalopoulos AS, Falagas ME. 2011. Colistin: recent data on pharmacodynamics properties and clinical efficacy in critically ill patients. Ann Intensive Care 1:30. doi: 10.1186/2110-5820-1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaye KS, Marchaim D, Thamlikitkul V, Carmeli Y, Chiu C-H, Daikos G, Dhar S, Durante-Mangoni E, Gikas A, Kotanidou A, et al. 2023. Colistin monotherapy versus combination therapy for carbapenem-resistant organisms. NEJM Evid 2:evidoa2200131. doi: 10.1056/evidoa2200131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. EUCAST . 2022. European committee on antimicrobial susceptibility testing. Breakpoint committee consultation on colistin October 2021. Colistin breakpoints - guidance document 2022
- 71. International Organization for Standardization . ISO. 2019. ISO 20776-1:2019.
- 72. European Committee on Antimicrobial Susceptibility Testing . 2020. Guidance document on broth microdilution testing of cefiderocol
- 73. European Committee on Antimicrobial Susceptibility Testing . 2024. Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST, version 14.0, 2024
- 74. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Matsen FA, Kodner RB, Armbrust EV. 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. 2012. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28:2223–2230. doi: 10.1093/bioinformatics/bts429 [DOI] [PubMed] [Google Scholar]
- 77. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, et al. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. DeTora LM, Toroser D, Sykes A, Vanderlinden C, Plunkett FJ, Lane T, Hanekamp E, Dormer L, DiBiasi F, Bridges D, Baltzer L, Citrome L. 2022. Good publication practice (GPP) guidelines for company-sponsored biomedical research: 2022 update. Ann Intern Med 175:1298–1304. doi: 10.7326/M22-1460 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1; Tables S1 to S11.
Data Availability Statement
Whole-genome sequencing variants were defined using the Bacterial Antimicrobial Resistance Reference Gene Database from the National Center for Biotechnology Information (Bioproject number: PRJNA313047). Data are available from Shionogi B.V. upon reasonable request.