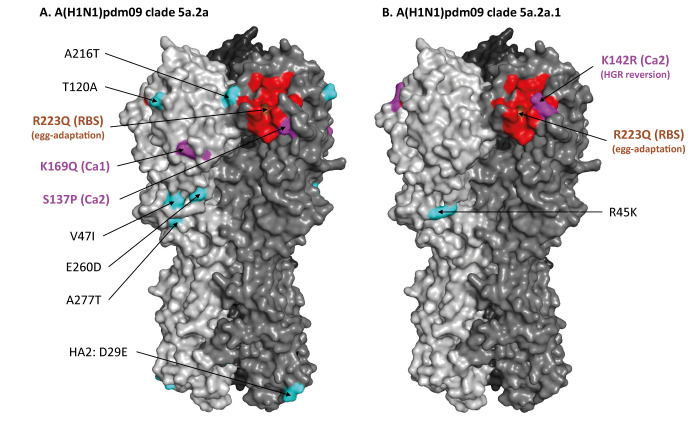
Figure 2.
Haemagglutinin substitutions present in circulating influenza A(H1N1)pdm09 clade 5a.2a and 5a.2a.1 viruses detected by the Canadian Sentinel Practitioner Surveillance Network, relative to the egg-adapted high-growth reassortant vaccine strain IVR-238, 2023/24 influenza season
HGR: high-growth reassortant; RBS: receptor binding site.
Representative influenza A(H1N1)pdm09 clade 5a.2a and 5a.2a.1 viruses were modelled in SWISS-MODEL [32] on A/Michigan/45/2015 (PDB 7knv.1) with substitutions highlighted in PyMOL relative to the egg-adapted HGR vaccine strain A/Victoria/4897/2022 IVR-238 [33]. The clade 5a.2a model shown in panel A depicts a combination of dominant T120A genetic clusters (Supplementary Table S2) rather than a single representative strain. The RBS is shown in red. Substitutions within the RBS are shaded brown. Substitutions in antigenic sites are shown in magenta and in cyan if non-antigenic. Amino acid numbering is based on A(H1)pdm09 numbering with the signal peptide removed. Vaccine-associated (vs circulating virus-associated) substitutions include Q223R, due to egg adaptation, and K142R, due to reversion in the HGR. Differences at position 216 may have an impact on glycosylation status as elaborated in the manuscript. Clade 5a.2a substitutions S137P (Ca2), E260D, A277T and HA2:D29E are clade-defining while other substitutions are new to one or more circulating 5a.2a subclades detected by the SPSN. The 5a.2a substitutions I96T (new) and HA2:H124N (clade-defining) are located inside the trimer interface and are therefore not visible in the structure.