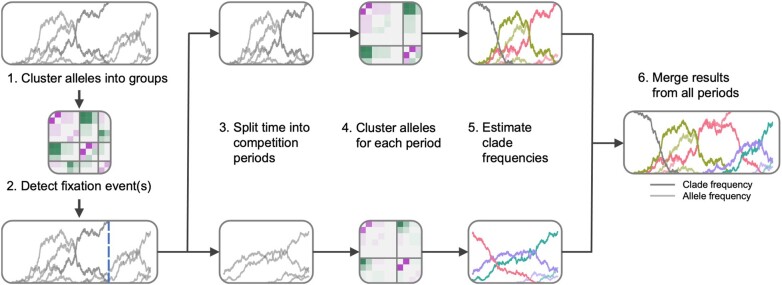
Fig. 1.
An overview of the method. We first compute the D matrix and cluster alleles into groups according to D matrix. The clustering results are reflected by the heatmap of the rearranged and segmented matrix D, where each block corresponds to a group. Aside from the group that consists of shared alleles which may or may not exist, each group consists of alleles that collectively compete with other groups. We then detect fixation events of alleles that are not shared. Once detected, we split time into competition periods at the fixation time(s). (n fixation events will result in competition periods.) We cluster alleles for each period. We then take the groups as initial clades and iteratively refine clonal identities and clade frequencies. Finally, we merge results from all periods into a complete reconstruction of the clonal interference throughout the evolution. The allele frequency trajectory data plotted in the figure are from a simulation, where a population of 1,000 sequences were simulated to evolve for 1,000 generations.