Fig. 8.
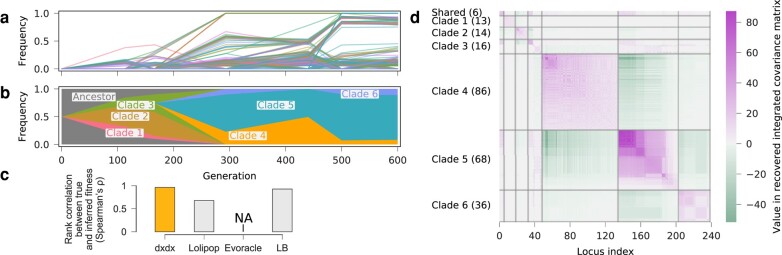
Clonal structure and fitness inference for B1 replicate of P. aeruginosa evolution data. a) Mutant allele frequency trajectories and b) clade frequency trajectories inferred by our method. c) We find a strong correlation between our fitness estimates and experimental measurements, exceeding those from Lolipop and similar to results following the LB method. Applying Evoracle to this data set yielded NaN results. d) The recovered integrated covariance matrix segmented into blocks according to clustering results of our method. The number of alleles in each clade is shown in brackets. Alleles in the same clades tend to show cooperating behaviors, as indicated by positive entries. Alleles tend to show competing behaviors across clades, as indicated by negative entries.