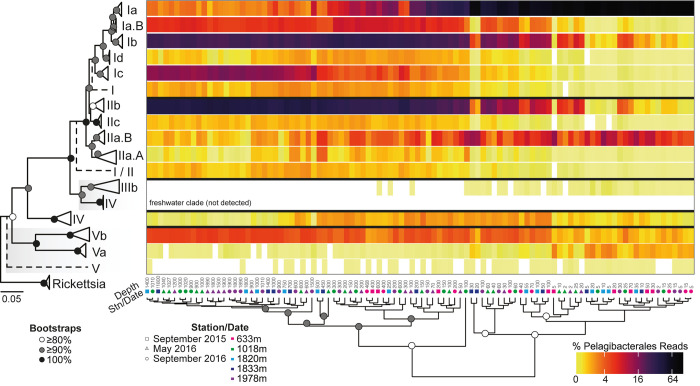
Fig 5. Phylogeny and distributions of Pelagibacterales.
The 16S rRNA gene phylogenetic reconstruction utilized full-length sequences and identifies 10 recognized subclades and 3 previously unrecognized subclades, all with bootstrap support >90% (1,000 replicates) except subclade IIb (86% support; see also S4 Fig). Note that dashed lines indicate LCA placements at unsupported nodes, some with relatively few amplicons. For example, that demarked ‘Clade V’—with a dashed line comprised >0.01% of amplicons . Some others with dashed lines were notable, but lacked full length sequences and therefore references sequences were not in the reconstruction. Also shown is the relative abundance of Pelagibacterales subclades as percent of all Pelagibacterales ASVs in each sample for the Monterey Bay Stations and depths sampled. Pelagibacter amplicons were first retrieved using PhyloAssigner and a global 16S rRNA gene reference tree [17]. These amplicons were then run in PhyloAssigner using the above Pelagibacterales reference tree. Heat map columns represent individual samples ordered by hierarchical clustering based on the Bray-Curtis similarities of the Pelagibacterales community composition. White in the heat map indicates not detected. Stations (color) and sampling dates (shape) are indicated as is sample depth (m) for each column.