Figure 6.
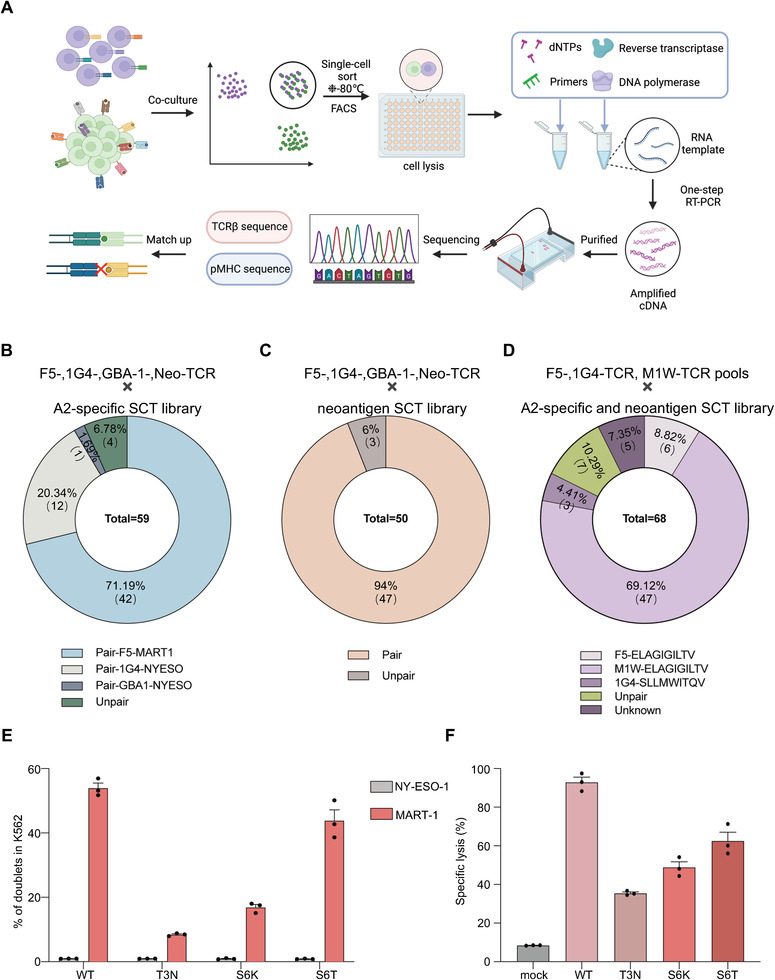
The library‐on‐library screening capability of DoubletSeeker. A) Schematic of DoubletSeeker for library‐on‐library screening of TCR‐pMHC interactions. Single doublets are sorted into individual wells of 96‐well plates, which are preloaded with cell lysis buffer, followed by freezing to lyse the doublet cells. Subsequently, the TCR and SCT information for each doublet is obtained through single‐cell PCR sequencing. B,C) The proportion of TCR‐pMHC pairs within the enriched doublets isolated from the co‐incubation of A2‐restricted SCT‐K562 library (B) or neoepitope SCT‐K562 library (C) with a mixture of F5‐, 1G4‐, GBA‐1‐ and Neo‐TCR‐Jurkat cells (TCR‐expressing cells: SCT‐expressing cells = 1:1). D) Proportion of TCR‐pMHC pairs within the enriched doublets isolated from the co‐incubation of the combined library of A2‐restricted SCT‐K562 and neoepitope SCT‐K562 with M1W‐TCR pools‐J76 cells, in the presence of a small proportion of F5‐ and 1G4‐TCR‐Jurkat cells (TCR‐expressing cells: SCT‐expressing cells = 1:1). E) Interaction between MART‐1‐K562 or NY‐ESO‐1‐K562 cells and J76 cells expressing M1WWT‐, M1WT3N‐, M1WS6K‐, or M1WS6T‐TCR as detected by DoubletSeeker. F) Cytotoxicity of T cells expressing M1WWT‐, M1WT3N‐, M1WS6K‐, or M1WS6T‐TCR against A2‐expressing K562 cells pulsed with MART‐1 peptide (ELAGIGILTV). The data are presented as percentage‐specific lysis rates. Data are represented as the means ± SEM. n = 3 (E and F).
