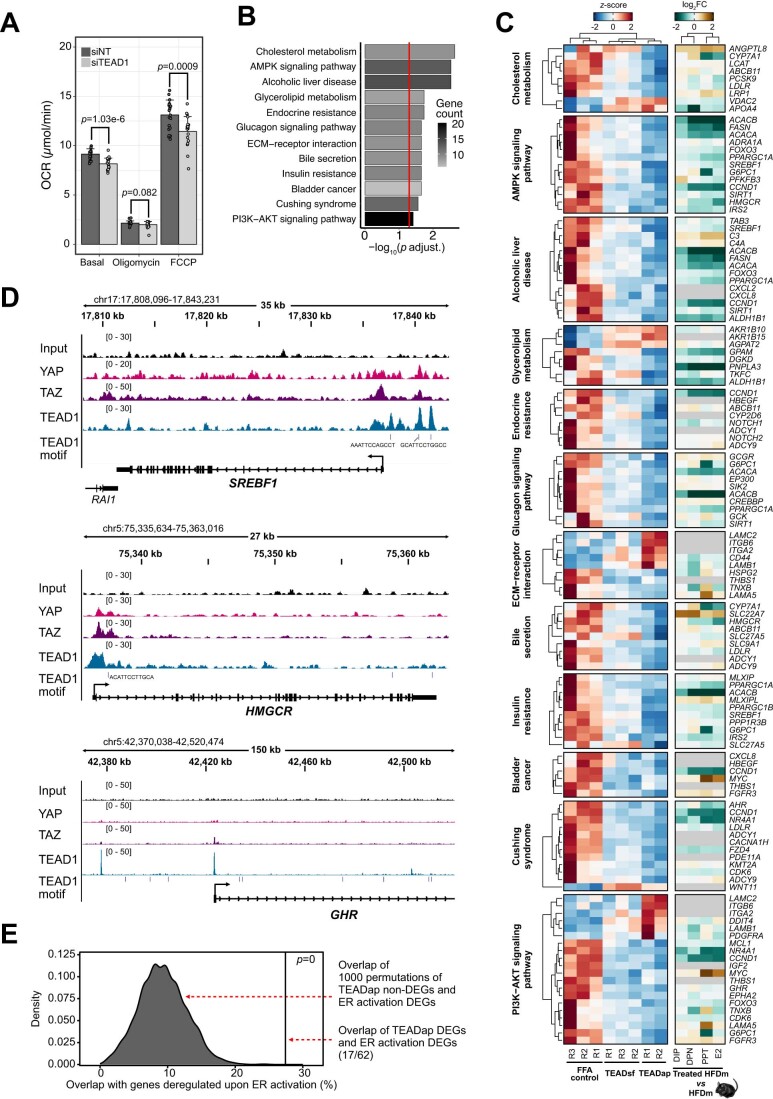
Figure EV5. TEAD inhibition changes key cellular processes in the liver.
(A) Bar plot displays mean of basal, minimum, and maximum oxygen consumption rate in HepG2 cells with control (siNT, dark gray) and siRNA-mediated TEAD1-KD (siTEAD1, light gray) (n = 63 per condition, +SD). Dots indicate single wells across two biological replicates. Significant P values are indicated (two-sided t test). (B) Bar plot shows the adjusted P values (log10) of the top 12 KEGG pathways enriched for genes deregulated upon TEADap treatment. Red vertical line indicates adjusted P value threshold (P < 0.05). Color gradient indicates gene number per pathway (low: gray, high: black). One-sided Fisher’s exact test with Benjamini–Hochberg correction was used for the overrepresentation analysis. (C) Heatmap displays gene expression changes of the top 12 KEGG pathways altered upon TEADap treatment. Left panel indicates normalized gene expression (z-score) in free fatty acid-fed controls, TEADsf and TEADap treatments (low: blue, high: red). Right panel shows log2FC for ER-agonist treatments compared to untreated male mice on HFD (low: green, high: brown). Gene names follow human nomenclature. Ortholog absence in mouse is specified (gray). (D) Genome browser views (IGV, hg38) show genomic regions around SREBF1, HMGCR and GHR gene loci. Genomic locations and sizes are indicated. The y axis of each track specifies normalized ChIP-seq read density (parenthesis) of YAP (pink), TAZ (purple) or TEAD1 (blue) in HUVEC cells (GSE163458). Vertical lines indicate TEAD1-binding sites identified by motif search. Blue boxes represent exons and UTRs, connecting lines indicate intronic sequences. Arrows indicate directionality of gene transcription. (E) Density plot shows the distribution of overlaps between 1000 random sets of genes unchanged by TEADap treatment and genes changed by ER-agonist treatments falling within the top 12-enriched KEGG pathways (gray peak). The percentage of overlapping genes changed in both TEADap and ER-agonist treatments for the top 12-enriched KEGG pathways is displayed (black vertical line). P value is indicated (permutation tests).